Molecules such as glucose, proteins, and other compounds can be ideal markers for detecting diseases such as diabetes due to their inherent biophotonic characteristics. However, the nature of the source of the sample (e.g., blood, urine, saliva) can introduce interference since some sample components can have similar photochemical properties when exposed to light with the same wavelength. Hence, the accuracy of results may not be consistent and reliable. Saliva should have fewer interfering proteins than blood-plasma samples with respect to biophotonic fluorescence. Thus, in these experiments, we have used saliva samples from 23 patients of different gender and age. Each saliva sample was mixed with a specific sensor constructed from DNA useful for fluorescence detection of glucose levels. The intensity of the fluorescence of each sample was compared to results from conventional clinical glycemic testing. The DNA sensor was developed based on synthetic biology, using a combination of synthetic and natural genetic parts: SNF3 + CYC1 Terminator + GAL1 Promoter + OGT + CYC1 Terminator + GAL1 Promoter + OGlcNA + CYC1 Terminator + GAL1 Promoter + EGFP. One of the advantages of this sensor is believed to be due to the presence of a specific gene related to a glycoprotein and/or protein modified with glucose found in high concentrations in saliva. The quantification of fluorescence measured using a photonics device showed greater sensitivity within a much shorter timeframe than the conventional glycemic techniques. Statistical analysis was carried out to enable specific identification, categorization, and quantification of the influence of different factors such as sample type (i.e. saliva), age, and gender of patient. Patients were grouped in different categories by integrating the different variables and were diagnosed with diabetes, pre-diabetes, or normal health based on predictions using the photonic data. Our investigation shows the advantage of using a non-invasive method such as a saliva test as well as a new highly sensitive detection method due to the interaction between the DNA-based sensor and the pho-tonicity.
DNA glucose sensor, diabetes, saliva, photon, fluorescence, synthetic biology
Diabetes is a degenerative disease caused by abnormal levels of glucose in the cell. These abnormal levels of glucose are not easily predictable and/or measurable [1]. Blood testing is the most common method to diagnose diabetes, however, this can be invasive and sometimes painful as well as potentially leading to microbial infection at needle insertion sites [2].
Current home use technologies determining glucose levels in the blood are not always accurate and may not be able to determine extremely low levels of glucose (below 20 mg/dL). Therefore, saliva could be an ideal substrate for determining glucose levels for reliable diabetes diagnosis [3]. However, the use of saliva for diabetes diagnosis is still very limited [4].
Most commercially available glucose meters for determining blood glucose levels in blood and plasma include enzymes such as glucose oxidase followed by measuring electric current produced during the enzymatic reaction. However, falsely high readings can occur depending on environmental conditions, which affect the efficacy of the enzyme and/or on specific medical treatments a patient is undergoing. Also, use of home portable, digital devices for monitoring glucose are known today. Although these digital devices produce quick results, the range of sensitivity can still be enhanced [5,6]. Therefore, molecular glucose sensors based on DNA have been previously developed by the main author of the present paper [7] in order to enhance sensitivity for detection of glucose based on fluorescence emission following interaction of a DNA-based glucose sensor with saliva. Furthermore, the sensitivity of the DNA glucose sensor is enhanced by the additional inclusion of signaling and/or transport specific glycoproteins or proteins modified with glucose genes [8,9].
Construction of DNA glucose sensor
The genes described below were assembled in pYES2 plasmid vectors by Clonetex Systems, Inc. (Austin, TX) and included an array of promoters and terminators. The full sequence of the DNA-based glucose sensor comprises an arrangement of genes, Gal1 promoters, and CYC1 terminators in the following order: SNF3 + CYC1 Terminator + GAL1 Promoter + OGT + CYC1 Terminator + GAL1 Promoter + OGlcNA + CYC1 Terminator + GAL1 Promoter + EGFP.
Sequences included the SNF3 glucose receptor with Genbank accession number XM_001386379.1 (a high affinity glucose receptor already proven as a glucose sensor), O-linked acetylglucosamine transferase (OGT) with Genbank accession number EAX05286.1, O-GlcNAc selective N-Acetyl-beta-D-glucosaminidase (O-GlcNAc) having Genbank accession number NP_571979.1, and Enhanced Green Fluorescent Protein (EGFP) having Genbank accession number ACV20892.1 (23). The glucose sensors were constructed by site-directed cloning with specific primers, as follows.
Assembly of glucose device
Procedure: This procedure was carried-out by combining standard experimental labora-tory methods of molecular biology and synthetic biology [7] with proprietary methods from Clonetex Systems, Inc. (Austin, TX). Thus, PCR amplification was per-formed as follows: Each gene was PCR amplified using gene-specific overlap primers and assembled sequences were subcloned into a HingIII- and XbaI-digested pYES2 vector.
PCR amplified pieces of all fragments were combined by using homologous recombination technology (Gibson Assembly). Clones obtained after transformation were sequenced and analyzed for DNA accuracy.
Design of glucose device: This device contains four genes and regulatory elements that are arranged in following order
SNF3 + CYC1 Terminator + GAL1 Promoter + OGT + CYC1 Terminator + GAL1 Promoter + OGlcNA + CYC1 Terminator + GAL1 Promoter + EGFP
At the completion of PCR of all four targeted genes, homologous recombination of the glucose detection device, and sub-cloning into pYES2 vector, four clones were select-ed from a transformed plate and processed for full length DNA sequencing. A clone with 100% matching sequence corresponding to the construct design was selected and purified to obtain plasmid construct at a mid-scale purification level. This DNA de-vice will allow the detection of proteins related to the presence of glucose in patient saliva samples as well as to detect existence of glucose itself.
Transformation of Yeast Saccharomyces cerevisiae (Sc) produced by Invitrogen, (INVSc1): The pYES2 plasmid assembled as described previously was transformed in-to competent S. cerevisiae cells following protocol provided by Invitrogen, Inc. (CA).
Production of glucose extract from yeast device
Transformed S. cerevisiae cells were grown in yeast malt broth at 30°C for 24 hours with an optimum OD of 2.56. An extract was produced from the yeast culture as follows: fermentation was conducted in yeast malt broth mixed with raffinose and galactose for 30°C for 72 hours. The cultures were then centrifuged at 9,000 rpm for 12 min at 15°C. Pellets were resuspended in 50 mL of sterile deionized water per gram of pellet. The resulting solutions were sonicated 4 times for a total of 2 minutes and 30 seconds and were then again centrifuged at 9,000 rpm for 12 min at 15°C. Supernatants were decanted and filtered with 0.56 μm filters and stored for later use.
In vivo fluorescence detection of DNA glucose sensor in saliva samples from patients
Preparation of samples: Saliva samples from fasting patients were collected by having the patients spit in a 3 cm × 2 cm collection tube until a total saliva volume of 3 mL per patient was collected. These samples were mixed with the DNA-based glucose sensors prepared as described above, at different ratios, and subjected to vortexing for 30-50 seconds. Although different ratios of the yeast DNA-based glucose sensors to saliva were used (i.e., 1:1, 2:1, 3:1, 6.3:1), the ratio 6.3:1 showed the best efficacy for each sample (i.e., 190μL of extract: 30μL of saliva).
Determination of fluorescence (FSU): A total volume of 200μL of saliva alone, DNA-based glucose sensor (DGS) alone, or DGS + saliva was used for fluorescence detection. The detection was performed using a Glomax E-8032: Multi+ Detection System with Instinct™ Software: Base Instrument with Shaking (PROMEGA, Madison, Wisconsin USA) by placing the selected sample on a plate reader, and the samples were exposed to a light source using the blue module (Ex: 490nm, Em: 510-570nm). Other excitation wavelengths were tested; however the selected blue light module showed the best consistent results. Fluorescence was determined based on fluorescence units (FSU), and compared to the results of the conventional glycemia tests. The natural inherent fluorescence (1,900 FSU) of the yeast was subtracted from the final fluorescence of each sample.
The fluorescence results were then divided in three groups according to the intensity of the fluorescence in relation to the clinical glycaemia concentration as described above [7] with the groups based on the means from ranges of fluorescence intensity. Samples were collected from 23 patients.
In vivo clinical glycemia determinations in blood plasma from patients
The clinical analysis of glycemia was performed at CONFA and Diagnostimed S.A Clinic laboratories (Manizales, Caldas, Colombia) during August-October of 2018. Glycemia was determined in blood from diabetics and non-diabetic patients by quantitative determination of glucose IVD using a standard colorimetric technique and Mindray BS380 equipment.
The results from the fluorescence analysis of the prepared sample with the DGS ex-tract and the results from clinical analysis of glycemia were then compared to the standard glucose value established by the Association of American Diabetics [10], shown in Table 1.
Table 1. Standard range of glycemia according AAD
Group |
Glycemia (mg/dL) |
Diabetic |
More than 126 mg/dL |
Pre-Diabetic |
Between 100 mg/dL and 125 mg/dL |
Healthy |
Less 100 mg/dL |
Statistical analysis
The statistical data was analyzed using Microsoft Excel 2013. An analysis of variance (ANOVA) was performed to determine the equal value of the means in the groups of patients and a linear regression was carried out in order to relate the study variables, in this case glycemia and fluorescence (extract + saliva of patients)
Construction of DNA glucose sensor
Assembly of the denoted DNA glucose sensor was effectively achieved, following standard molecular biology and synthetic biology procedures [7]. Thus, the different genetic parts of the sensor, which include Gal1 promoter and CYC1 terminator, along with the other genes SNF3, OGT, OGlcNA, and EGFP were fully integrated and sub-cloned into PYES2 vector (Figure 1). Then, four clones were selected from transformed plates and processed for full length DNA sequencing (sequencing data is provided electronically). Clones with 100% DNA sequence accuracy were selected for further processing. The assembly of the construct DNA glucose sensor was also confirmed by PCR analysis, which showed full assembly of the different genetic parts [7].
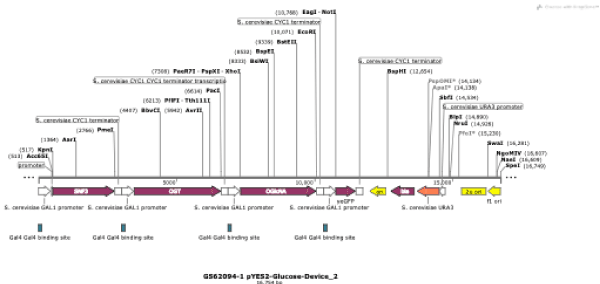
Figure 1. Linear map of a plasmid containing the genetic elements of the DNA-based glucose sensor for detecting glucose in saliva
Selection of positive yeast clones transformed with the DNA-based glucose sensor
Yeast competent cells (strain INVSc1 from Invitrogen) were transformed with the recombinant plasmid discussed previously and selected on synthetic complete (SC) dropout plate (deficient in uracil base). A well-isolated clone was picked from the SC plate.
In vitro analysis of the DNA-based glucose sensor in saliva samples from patients
Results show higher fluorescence intensity (13,000 FSU) in saliva samples, which directly correlates with higher glycemia levels in patients previously diagnosed with diabetes (161 mg/dL). Furthermore, saliva samples with low fluorescence (7,000 FSU) corresponded to samples from healthy patients with lower glycemia level (65 mg/dL). Finally, a middle range of fluorescence (10,500 FSU) corresponded to medium glycemic levels (123 mg/dL) in patients diagnosed as pre-diabetic (Table 2).
Table 2. Results of fluorescence intensity and glycemia of patients placed in the following groups
Patient Age |
Gender |
DNAGYE + Saliva Initial Fluorescence (FSU) |
DNAGYE + Saliva Final Fluorescence (FSU) |
Clinical Glycemia (mg/dL) |
|
Group 1: Diabetics Mean= 6957.143 +/- 822.3 |
36 |
Female |
10600 |
6000 |
127 |
38 |
Female |
11000 |
6400 |
135 |
39 |
Female |
11200 |
6600 |
129 |
31 |
Female |
11300 |
6700 |
132 |
30 |
Male |
11500 |
6900 |
172 |
35 |
Female |
12300 |
7700 |
152 |
44 |
Female |
13000 |
8400 |
161 |
|
Group 2: Pre-Diabetics. Mean = 5500 +/- 308.22 |
23 |
Male |
9700 |
5100 |
117 |
42 |
Male |
10000 |
5400 |
102 |
40 |
Female |
10000 |
5400 |
123 |
37 |
Male |
10300 |
5700 |
125 |
45 |
Male |
10500 |
5900 |
123 |
|
Group 3: Healthy Mean = 3600 +/- 930.6 |
4 |
Female |
7000 |
2400 |
65 |
18 |
Female |
7300 |
2700 |
77 |
1 |
Female |
7500 |
2900 |
84 |
12 |
Female |
7500 |
2900 |
84 |
3 |
Female |
7600 |
3000 |
73 |
14 |
Female |
7700 |
3100 |
77 |
28 |
Female |
8600 |
4000 |
84 |
24 |
Female |
9000 |
4400 |
89 |
43 |
Female |
9000 |
4400 |
93 |
6 |
Female |
9500 |
4900 |
97 |
25 |
Female |
9500 |
4900 |
97 |
Group 1=Diagnosed diabetics, Group 2=Pre-Diabetics, and Group 3=Healthy
Note 1. DNAGYE: DNA Glucose Yeast Extract, FSU: Fluorescence units
*”DNAGYE+Saliva Final Fluorescence (FSU)” is the fluorescence background corrected for extract fluorescence (4600 FSU)
Note 2. Results are based on the mean of number of patients per group, after ANOVA.
Note 3. p-value=1.5×10-7
Statistical analysis allowed us to use the saliva results from patients in order to group them in three categories, Group 1=Diagnosed Diabetics, Group 2=Pre-Diabetics, and Group 3=Healthy. Significant differences were found among the groups (P <0.05) (Table 3) a relationship between fluorescence and glycemia was also discovered, as shown in Table 2 and Figure 2.
Table 3. Ranges of fluorescence intensity and glycemia levels according the patient groups 1, 2, and 3
Group |
Range of glycemia |
Range of Fluorescence (FSU) |
Diabetic |
>126 mg/dL |
10,600-13,000 |
Pre-Diabetic |
100 mg/dL-125 mg/dL |
9,700-10,500 |
Healthy |
<100 mg/dL |
6,000-9,500 |
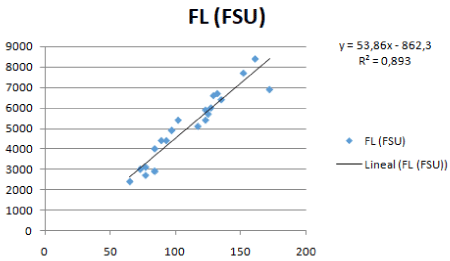
Figure 2. Regression analysis between fluorescence and glycemia
Ranges of fluorescence correlated with ranges of clinical glycemia, which are provided in Table 3.
Results showed the efficacy of assembling the DNA-based glucose sensor sub-cloned in yeast to detect glucose in saliva using molecular and synthetic biology techniques. Previous reports have shown similar results in blood plasma (Cuero et al 2017). The current results corroborate the efficacy not only of the assembly of the different genetic parts of the DNA-based glucose sensor, but also the positive effect of the two specific post-transcriptional proteins OGT and OGlcNAc on determining glucose levels in saliva.
The advantage of these two proteins seems to be based on their effective complemen-tary catalytic processes, in which O-GlcNAc transferase (OGT) is responsible for cata-lyzing the addition of O-GlcNAc to serine/threonine residues, while β-D-N-acetylglucosaminidase (O-GlcNAcase) catalyzes removal of O-GlcNAc. OGT appears to bind to mediating transporters of sugars such as UDP-GlcNAc, which affects proteins related to glucose production. Furthermore, OGT transfers the N-acetylglucosamine (GlcNAc) moiety onto serine/threonine residues of target proteins related to glucose metabolism [8]. Consequently, higher amounts of glucose in the saliva sample would result in higher activity levels for these two proteins. These two proteins have previously been detected in different tissues including muscle, liver, and kidney [8]. However, the current research represents one of the first reports using saliva as a substrate for these two proteins assembled as part of a DNA-based glucose sensor.
The present investigation shows that saliva can be an ideal substrate for early detection of glucose at extremely high or low concentrations. The increased sensitivity is perhaps due to fewer potentially interfering molecules (e.g. proteins) present in blood, which can introduce interference with detection. Early investigations in blood using the DNA-based glucose sensors described herein in combination with fluorescence detection corroborates lower sensitivity when using blood as a substrate [7]. Currently, the author of this paper is enhancing the sensitivity of this technology using saliva.
Saliva seems to be a good substrate for early detection of glucose levels. The high sensitivity of the present technology is enhanced by the specific genes and proteins (i.e. OGT and OClcNA) used in the construction of the DNA-based glucose sensor.
This technology could be used alone and/or as a complementary technique to many existing technologies, which can provide quick detection but not necessarily good sensitivity. For example, in enriching analysis of otherwise non-detectable extreme concentrations (i.e. maximum or minimum) technology described is able to analyze samples with glucose concentrations outside of standard clinical ranges.
This work was supported by the International Park of Creativity laboratory (www.ipoc.co), Manizales, Colombia, and BioCapital Holdings, LLC, USA (www.biocapitalholdings.com). We thank CONFA and Diagnostimed S.A Clinic Laboratories (Manizales, Colombia) for the clinical analysis of glycemia, and Clonetex Systems Inc., Austin, TX, for providing the synthesized sequences as well as for technical assistance during the implementation of this work. Also, we thank Dr. Lawrence Villanueva for editing the manuscript.
- Singh S (2011) The genetics of type 2 diabetes mellitus: A review. J Sci Res 55: 35-48.
- Smriti K, Pai KM, Ravindranath V, Gadicherla S, Pentapati KC (2016) Salivary glucose as a diagnostic marker for diabetes mellitus. J Diabetes Sci Technol 10: 991-992. [Crossref]
- Streckfus CF, Bigler LR (2002) Saliva as a diagnostic fluid. Oral Dis 8: 69-76. [Crossref]
- Gupta S, Meghanand TN, Sunitha JD, Dawar G, Sinhs, et al. (2017) Correlation of salivary glucose level with blood glucose level in diabetes mellitus. Journal of Oral and Maxilofacial Pathology 21: 334-339. [Crossref]
- Frias JP, Lim CG, Ellison JM, Montandon CM (2010) Review of adverse events associated with false glucose readings measured by GDH-PQQ-based glucose test strips in the presence of interfering sugars. Diabetes Care 33: 728-729. [Crossref]
- Kelly BN, Haverstick DM, Bruns DE (2010) Interference in a glucose dehydrogenase- based glucose meter. Clin Chem 56: 1038-1040. [Crossref]
- Cuero R, Naïve J, Agudelo D, Medina P (2017) Construct of DNA glucose sensor yeast plasmid for early detection of diabetes. Integrative Obesity and Diabetes 3: 1-9.
- Junfeng M, Hart G (2013) Protein O-GlcNAcylation in diabetes and diabetic complications. National Institute of Health 10: 365-380.
- Slawson C, Copeland RJ, Hart GW (2010) O-GlcNAc signalling: A metabolic link between diabetes and cancer? Trends Biochem Sci 35: 547-555. [Crossref]
- American diabetes association (2002) Screening for diabetes. Diabetes care 25: 21-24.


