Background: In Israel during the first weeks of 2014, computerized HMO records indicated a rise in acute gastroenteritis among children < 2 yr with a concomitant increase in rotavirus-positive stools. Children (80%) received 3 doses of pentavalent rotavirus vaccine since 2011. Retrospective analysis showed a vaccine effectiveness of 70% for pediatric RAGE cases between September 2013 and January 2014.
Aim: To rule in or out the presence of a vaccine escape mutant.
Methods: Point prevalence genomic analysis (rapid genotyping of rotavirus from rotavirus–positive dipsticks collected from community cases between January 20 to 23, 2014) and continued syndrome surveillance.
Results: Twelve of 24 rotaviruses identified from 23 dipsticks (one was a double infection) were G12P[8] rotaviruses. The initial rapid rise in RAGE cases was not sustained.
Conclusion: The overall genetic pattern of isolated rotavirus was consistent with endemic circulation of different rotaviruses, reflecting the global emergence of G12 strains, rather than circulation of a specific emerging monophyletic genotype. This interpretation was strengthened by the subsequent falloff of additional RAGE cases. Conducting different surveillance strategies in parallel (in this case syndrome surveillance, laboratory surveillance, and rapid genetic characterization of isolates) strengthens the reliability of interpreting data from any one of the platforms especially when sample sizes are small and the ability to detect and characterize potential spreading events at early stages.
rotavirus, rotavirus genotyping, acute gastroenteritis, syndrome surveillance, rotavirus vaccine
Group A rotaviruses cause approximately 527,000 deaths in children under 5 years of age, mainly in developing countries [1,2]. The rotavirus genome consists of eleven double stranded RNA segments, ten of which encode a single viral protein, and one that encodes two proteins [3]. Two independent neutralization antigens, VP7 (which defines G-types) and VP4 (which defines P-types) reside on the outer protein layer of the virion [3,4].
All Israeli citizens must be enrolled by law in one of four Health Maintenance Organizations (HMOs). The Israel Center for Disease Control (ICDC) conducts weekly syndrome surveillance for acute gastroenteritis (AGE) based on screening ICD-9 codes for visits provided by the second largest HMO, covering approximately 25% of the population.
Two live rotavirus vaccines, RotaTeq (Merck) and Rotarix (Glaxo-SmithKline), have shown efficacy of 85%–98% against severe cases of acute gastroenteritis caused by rotavirus (RAGE) [5,6]. Rotarix contains rotavirus expressing G1 and P[8] antigens while Rotateq contains rotaviruses expressing G1, G2, G3, G4 and P[8] antigens. In Israel, both vaccines were available for purchase starting in 2008, until RotaTeq became free after December 2011 when it was added to the routine childhood vaccination schedule. Ministry of Health computerized vaccination records indicated 81.7%, 80.5%, and 78.5%, 3-dose vaccine coverage with Rotateq in 2011, 2012 and 2013, respectively. The effectiveness of the vaccines has been reported elsewhere [7-12]. On January 15, 2014, ICDC AGE surveillance indicated a high rate increase in the incidence of rotavirus positive stools in children < 2 years old than had been observed during the previous 2012-2013 season. To rule out emergence of a rotavirus that escaped protection by the RotaTeq vaccine, we rapidly determined the P- and G-genotypes of rotaviruses in stools sent to the Maccabi Health Care Services lab between Jan 20 and 23, 2014 as described by Shulman et al. [13] without increasing the workload of that primary diagnostic laboratory. This report describes the phylogenetic evidence obtained from G and P gene sequences of RNA extracted from the rotavirus-specific bands on the rotavirus-positive dipsticks set aside by the HMO laboratory combined with continuation of syndromic and laboratory surveillance that was used to rule out emergence of a new strain.
Ethics statement
The Ethical Review Board of the Sheba Medical Center, Tel Hashomer, approved this study (SMC-0217-13) and exempted it from a requirement to obtain informed consent. Data was stripped of all personal details pertaining to, or which could be used to identify rotavirus-positive individual patients. The HMO Central Laboratory provided the gender, age, and residential cities of all of the rotavirus positive patients.
Patient and clinical samples
Stool suspensions collected between January 20 and January 23, 2014 from 12 male and 11 female children (mean: 2.06 yr; median: 1 yr; with 18 of 23 < 2 yrs; range: 8 month to 7 years) tested positive for rotavirus at the Maccabi Health Care Services Central Laboratory using diagnostic rotavirus rapid test strips (dipsticks). With the exception of two patients, all children lived in different cities spread throughout north, central, and southern Israel.
Genotyping
Sequence grade RNA was extracted from excised rotavirus bands on the dipsticks as previously described [13]. cDNA was prepared with M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and random primers (Cat. no. 48190011, Invitrogen, Carlsbad, CA, USA) by denaturing the RNA (97°C for 5 mi, then on ice), adding 15 µl of reaction mix (1 µl RT enzyme, 7 µl 5x buffer, 3.5 µl 40 mM MgCl2, 0.7 µl random primers, 1 µl 10mM dNTP mix, 0.5 µl RNAse inhibitor (40 U/µl), and 1.3 µl sterile water) and incubating at 37°C for 60 min). The cDNA was amplified using AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA) and the generic external primers for P-typing and G-typing described by Gunasena et al. [14] and Gouvea et al. [15], respectively. P-typing: 2 µl of cDNA, was added to 48 µl of a mix containing 4.8 µl 10x Invitrogen Taq buffer, 2.5µl 50 mM MgCl2, 1 µl of 10 mM dNTP mix, 0.2 µl Invitrogen Taq polymerase, 7 µl primer mix, and 32 µl sterile water. Tubes were heated to 94°C for 5 min; then run 30 cycles of 1 min at 94°C, 2 min at 45°C, and 1 min at 72°C, followed by a final elongation at 72°C for 7 min. G-typing: 2 µl of cDNA was added to 48 µl of a mix containing 4.8 µl 10x Invitrogen Taq buffer, 2.5 µl 50 mM MgCl2, 1 µl of 10 mM dNTP mix, 0.2 µl Invitrogen Taq polymerase, 9 µl primer mix, and 35.3 µl sterile water; heat to 94°C for 5 min. Tubes were heated to 94°C for 5 min, run through 30 cycles of 1 min at 94°C, 2 min at 42°C, and 1 min at 72°C, followed by elongation at 72°C for 7 min.
Sequence analysis of both strands was as previously described [13]. Genotypes were identified by comparison with sequences of rotavirus prototypes using the Sequencher program v5.2 (Genecodes, Anne Arbor, Michigan, USA), BLAST to search the DDBJ/EMBL/GenBank sequence database [16], and RotaC automated genotyping tool for Group A rotaviruses [17]. The naming convention for the Israeli sequences is: RV_patient number_Israel2014_G-typeP-5type. The sequences have been deposited the DDBJ/EMBL/GenBank sequence database (Access numbers MH357640 to MH357681).
Phylogenetic analysis
VP4 and VP7 sequences were aligned on Sequencher v5.2 and trimmed to the length of the shortest sequence that would include the most sequences from each genotype; 518, 337, and 431 nt to include 9 of 10 G3 sequences, 8 of 12 G12 sequences and 21 of 23 P[8] sequences, respectively. Separate neighbor-joining trees were constructed using Clustal [18] for each of the three genes after bootstrapping data 1000 times. Phylogenetic trees were visualized using NJplot v2.3 http://pbil.univ-lyon1.fr/software/njplot.html (last accessed Oct 2017).
Based on a higher and more rapid increase in the number of rotavirus-positive stools in children < 2 years at the start of the winter rotavirus season in 2014 (Fig 1, black arrow and lower curve), compared with 2013, it was decided to determine the point prevalence of rotavirus genotypes in relation to the genotypes of the Rotateq and RotaRix vaccines in use in Israel between 2008 to 2014 or to 2011, respectively, to rule out emergence of a strain resistant to the protection provided by the vaccine strains. The incidence of all visits for gastroenteritis is represented by the upper curve in figure 1.
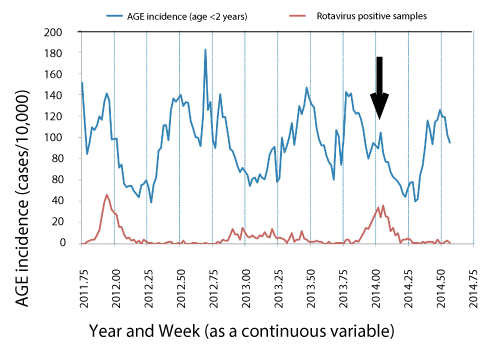
Figure 1. Syndrome surveillance for acute gastroenteritis in children < 2 yr in the community (Panel A) compared with the weekly number of rotavirus antigen positive stools up to week 2 (Panel B) and week 30 (Panel C) of 2014. The upper trace indicates the weekly number of visits for acute gastroenteritis in children < 2-year-old presenting at the community Maccabi Health Care Services per 10,000 children reported up to week 30 of 2014. The lower trace indicates the weekly number of laboratories confirmed rotavirus-positive stools (Maccabi Health Care Services Central Laboratory). The solid black arrow indicates the date at which the rapid upward trend in acute gastro enteritis and rotavirus positive stools triggered the request for genetic analysis of circulating rotavirus strains. The figure is a composite graph from weekly reports prepared by the Israel Center for Disease Control and is presented here with their permission
Rotavirus RNA was extracted from the rotavirus-positive band on diagnostic strips from all 23 rotavirus-positive stool tested by the Maccabi Health Care Services Central Laboratory between January 20th and January 23rd, 2014. The P- and G-genotypes were determined as described in the methods. The distribution of the genotypes is shown in table 1. The stool sample from patient 4 contained a mixture of two different G-types with a single P-type. Nucleotides mixtures at 5 nucleotide positions in a single G-type in stools from patient 22, indicated a homotypic mixture of different G12 genotypes.
Separate neighbor-joining trees were constructed using Clustal X for P[8] and for G3, G12 sequences after bootstrapping data 1000 times (Figure 2, left and right sides, respectively). The name of each sequence on each of the phylogenetic trees lists both the P and G-type. BLAST searches of representative sequences from each major branch indicated matches at > 98% homology with contemporary (2011-2014) isolates from India, the Far East, Africa and Europe as well to much earlier isolates (not shown).
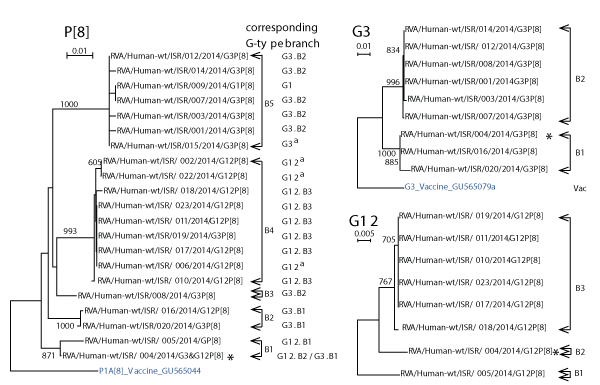
Figure 2. Phylogenetic Neighbor-joining trees of P[8], G3 and G12 sequences of RNA isolated from dipsticks collected between January 20th and 23rd.
Neighbor joining trees were prepared for Israeli G3, G12, and P[8] sequences for sequences of length 518, 337, and 431, respectively. The sequences were bootstrapped 1000 times. The numbers at branch points indicate bootstrap values > 75%. Brackets to the right of each phylogenetic tree indicate separate branches containing closely related sequences. Each branch has been assigned an arbitrary branch name beginning with the letter “B” followed by the arbitrary number of the branch. The G-types and its assigned branch names are also listed in a column to the right of each sequence in the P[8] phylogenetic tree to identify the G-type associated with that P-type in the clinical sample. A G-type followed by a superscript “a” instead of a G-type branch number indicates that the G-type sequence was sufficiently long to identify the G-genotype but insufficiently long to appear in the respective G3 and G12 phylogenetic tree. An asterisk was placed to the right of G-types and P type of patient 004 to indicate that stools from this patient contained a mixture of two rotaviruses with different G-types and a single P-type.
Table 1. Distribution of Rotavirus Genotypes isolated between January 20 to January 23, 2014.
Genotype |
Number |
G1P[8] |
2 |
G3P8] |
10 |
G12P[8] |
12 |
Total |
24 |
G3 isolate sequences segregated into two branches (B1-B2 in figure 2, top right). The G3s varied by 37 (7%) to 47 (9%) out of 518 nucleotides from the G3 RotaTeq vaccine strain sequence.
G12 isolate sequences segregated into 3 branches (B1-B3 in figure 2, bottom right). The G12 sequences were compared with the four lineages of G12 reported by Matthijnssens, et al. [19] and Rahmanet al. [20] (Figure 3). All Israeli sequences belonged to lineage GIII except for sample 005 that was distant from representatives of all four lineages. Aoki et al. [21] described amino acid residues in the VP7 gene product of G3 rotaviruses that defined antigenic epitopes 7-1a; 7-1b; and 7-2 and CA+2 binding sites. G12 nucleotide sequences for patients 005 (Branch B1, Figure 2); 010, 011, 017, 019, and 023 (Branch B3, Figure 2), and a G12 (DQ146665) from Branch GIII in figure 3, were translated in silico using Sequencher. The amino acid sequences were aligned with amino acid sequences from a G3 (AB091777) isolate to identify equivalent residues from antigenic and Ca+2 binding sites. The Israeli G12 sequences differed from G3 at seven, three and two amino acid residues in antigenic epitopes 7-1a; 7-1b; and 7-2 described for G3 by Aoki et al. [21] (Table 2). Among the Israeli G12 sequences all except one amino acid residue in isolate 005 in epitope 7-1a were identical despite large nucleotide differences. All Ca+2 binding amino acid residues were conserved. Without testing, it is not possible to predict the effect of amino acid differences in residues within and adjacent to the residues listed in table 2.
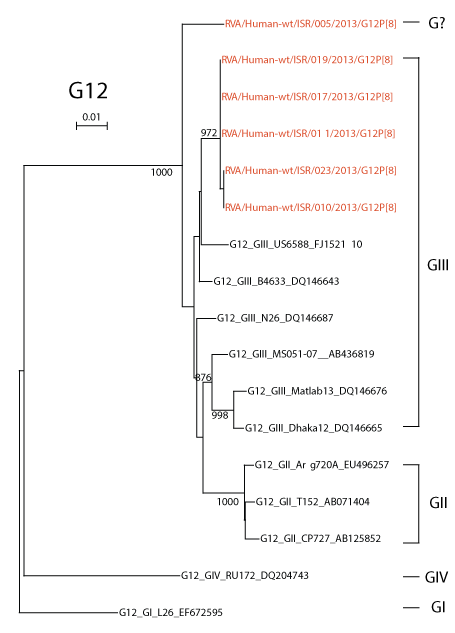
Figure 3. Phylogenetic Neighbor-joining trees of G12 sequences of RNA isolated from dipsticks compared with sequences representing different G12 lineages.
A neighbor joining tree was prepared with Israeli G12 sequences of 661 nt in length and sequences representative the four main G12 lineages reported by Matthijnssens, et al. [19] and Rahman et al. [20]. Numbers on branches indicate bootstrap values >75%. [Note: Two Israeli sequences RVA/Human-wt/ISR/004/2013/G12P[8] and RVA/Human-wt/ISR/018/2013/G12P[8] from Figure 2 were not included in this phylogenetic analysis since readable sequences were less than 661 nt in length.]
Table 2. Amino acid residues in antigenic epitopes and the Ca+2 binding site in G12 rotaviruses isolated in Israel in 2013 inferred from the antigenic epitopes and the Ca+2 binding site for G3 rotaviruses described by Aoki et al. [21]. An “=” means that the amino acid residue was not different from AB091777; A “?” indicates amino acid residues with tentative equivalency to the epitopes described in Aoki et al. [21].
Site |
Antigenic Epitope |
Residue (G3) |
G3
(AB091777) |
G12 GIII
(DQ14665) |
ISR G12 B1/? |
ISR G12
B3/GIII |
7-1A |
A |
87 |
T |
S |
S |
S |
7-1A |
A |
91 |
T |
N |
T |
T |
7-1A |
A |
94 |
G |
T |
T |
T |
7-1A |
A |
96 |
T |
P |
P |
P |
7-1A |
A |
97 |
E |
D |
D |
D |
7-1A |
A |
98 |
= |
= |
= |
= |
7-1A |
A |
99 |
K |
T |
T |
T |
7-1A |
A |
100 |
D |
N |
N |
S |
7-1A |
A? |
104 |
= |
= |
= |
= |
7-1A |
D |
291 |
= |
= |
= |
= |
7-1B |
E |
190 |
= |
= |
= |
= |
7-1B |
C |
201 |
= |
= |
= |
= |
7-1B |
C |
211 |
N |
D |
D |
D |
7-1B |
C |
213 |
A |
T |
T |
T |
7-1B |
C |
217 |
= |
= |
= |
= |
7-1B |
F |
238 |
D |
N |
N |
N |
7-2. |
B? |
145 |
D |
Q |
Q |
Q |
7-2. |
B |
147 |
T |
S |
S |
S |
7-2. |
B |
148 |
= |
= |
= |
= |
7-2. |
C |
221 |
S |
A |
A |
A |
7-2. |
C |
223 |
= |
= |
= |
= |
7-2. |
F |
242 |
= |
= |
= |
= |
7-2. |
B |
264 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
95 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
177 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
182 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
204 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
214 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
216 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
228 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
229 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
231 |
= |
= |
= |
= |
Ca+2 |
Ca+2 |
301 |
= |
= |
= |
= |
The P[8]s segregated into five different branches ((B1-B5 in figure 2, left). P[8] isolate sequences differed from the RotaTeq P[8] vaccine strain by 24 (7%) to 33 (10%) out of 341 nucleotides that were compared. The branch designations of each G-type taken from the G3 and G12 phylogenetic trees on the right in figure 2 are listed to the right of each isolate on the P[8] phylogenetic tree to make it easier to visualize the combination of G- and P-types in isolates from each patient.
Syndrome surveillance has been used as a means of identifying unusual or unexpected changes in rates of illness that may indicate the start of a natural event, an accidental event, or deliberate (bio-terror) event [22]. Syndrome surveillance is based on measuring changes in incidence of symptoms or behavior patterns caused by pathogens in real time before an unusual epidemiological event is recognized and diagnosed. As an unusual epidemiological event unfolds, people develop symptoms that lead to changes in behavior patterns (for example absence from work or school, increased purchases of over-the-counter palliative medicines), followed by visits to physicians where they first present with general symptoms and later with more specific symptoms. Unfortunately, most pathogens that trigger unusual events, share symptoms with events caused by endemic or “routine” pathogens, particularly at early stages of their activity making it difficult to recognize that they represent the start of an unusual epidemiological event.
The effectiveness of syndrome surveillance requires systematic observation of natural trends over prolonged periods of time. In the current case the triggering event for rapid response (determining the point prevalence of circulating rotavirus genotypes) was the sharp rise observed in acute gastroenteritis in children < 2 years that corresponded with a rise in rotavirus-positive stool samples among the general population of one HMO representing 25% of the Israeli population at the start of the 2013-2014 rotavirus winter season. The initial rate of increase for laboratory confirmed rotavirus cases resembled the rate of increase in pre-rotavirus vaccination periods. This rise could portend the start of circulation of a new strain able to evade protection from the newly introduced RotaTeq vaccine or merely reflect seasonal differences.
A 70% vaccine efficacy was estimated for Israeli children based on the vaccine history of pediatric rotavirus-positive stools reported by community clinics at the beginning of the 2013-2014 winter season and an 89% vaccine effectiveness (95% CI 51.9% to 97.6%) against severe rotavirus acute gastroenteritis requiring hospitalization of children 0 to 5 years old [7]. These are similar rates to those reported by Vesikari et al. [6] against any G1–G4 rotavirus gastroenteritis through the first full rotavirus season after vaccination with the pentavalent rotavirus vaccine (74.0 percent; 95% confidence interval, 66.8 to 79.9 percent).
In the primary HMO central diagnostic laboratory, rotavirus diagnosis by dipsticks is only requested for a subset of all stool samples sent for diagnostic testing. Moreover, a rotavirus-positive stool sample might be located at various other workstations since rotavirus testing is usually only one of a series of tests requested for the given sample. Providing original stool samples for sequencing in a sequencing laboratory carried the risk of laboratory contamination in the large automated primary laboratory by temporarily disrupting the SOP for stool diagnosis. It would also require a person to sort through large numbers of stool samples located at different workstations, additional facilities for subsequent longer-term storage of the samples, and provision for safe transportation of stool samples between labs. For these reasons, sequence analysis of rotavirus RNA from rotavirus-positive dipsticks [13] offered a much less disruptive method to characterize the rotavirus in the rotavirus positive stool samples. Positive dipsticks can be stored in easily transportable envelopes at room temperature for at least 5 years [13]. In the present study, air-dried rotavirus–positive dipsticks were collected at the end of each workday from the rotavirus testing station and storage at room temperature was only for a few days after collection. Thus, disruption was minimal and no special storage facilities or storage temperatures were required.
Sequence grade RNA was extracted from all dipsticks and the sequences were of enough quality and length for genotypic and phylogenetic analysis. The phylogenetic analysis was quite informative. The topography, multiple branches of contemporary sequences in the phylogenetic trees, was consistent with endemic transmission of multiple strains rather than exclusively to emergence of a single genotype. Comparison of P[8] and G3 sequences with respective RotaTeq vaccine sequences ruled out infection by vaccine or vaccine-derived strains. The samples were from patients living in different cities. Analysis of the pattern of association between the G-type and P-type in individual patients is consistent with endemic transmission since lineages of each G-type segregated together with a specific lineage of P-type. Patient 004 had a double infection as RNA from the dipstick encoded two different G –types and a single P-type.
Half of the rotaviruses sampled had a G12-genotype that is not directly covered by the G-types in the Rotateq vaccine. All G12 isolates had the P[8]-genotype which is present in the Rotateq and Rotarix vaccines. Both vaccines convey protection against severe infections (e.g. those that require hospitalization), even those caused by rotaviruses by G-types that were not included in the compositions of either vaccine [5,6,23].
G12 genotypes emerged in 1987 and rapidly spread throughout the world (reviewed in [19,20]). A single sub-lineage, GIII, was responsible for this global spread [19]. G12P[8] rotaviruses have been identified in Israel since 2007 in children hospitalized for RAGE ([8] and the Annual Reports of the Israel National Center for Viral Gastroenteritis). All of the rotaviruses reported in the present study were isolated during a four-day period in the middle of the 2013-2014 rotavirus season in Israel from children with less severe cases, e.g. cases that did not require hospitalization. G12P[8] was the predominant cause of their rotavirus acute gastroenteritis. There were no differences in amino acid residues in antigenic sites among the Israeli G12 isolates (with one exception for the isolate from patient 005) despite differences in nt sequences, implying that the G12 isolates had similar antigenic properties. There was also only a single amino acid difference with the GIII isolate (DQ14665, Table 2) used for comparison. Rotavirus acute gastroenteritis caused by G12P[8]s rose from 1% of hospitalized cases before adoption of the universal rotavirus vaccination program in Israel to 4.7% in the five years after adoption of universal vaccination [9,10]. This does not automatically mean that G12P[8] rotaviruses evaded the protection afforded by the rotavirus vaccine, since each rotavirus also contained P[8] genes. However, it is important to continue to monitor the genotypes of hospitalized and non-hospitalized cases to see whether the proportion of G12P[8] cases increase and whether similarities in amino acid residues in antigenic sites might be related to selective pressure by vaccination. The ratio of G12P[8]s in hospitalized cases to non-hospitalized cases would also provide an indication of any changes in severity of gastroenteritis caused by circulating G12P[8] isolates relative to the other co-circulating rotaviruses. With the exception of one G12P[8] isolate that did not link closely with representative sequences of any of the four major G12 lineages, all of the isolates from the less severe cases reported here belonged to the predominant GIII lineage. Thus, it is not surprising that the Israeli sequences were very closely related to sequences from many different regions of the globe. The interpretation that the G12 strains were not strains that escaped vaccine protection was born out by the fact that the initial rapid rise in AGE and in rotavirus positive stools was not sustained.
In conclusion, our study emphasizes the need for continued rotavirus surveillance in order to detect potential spreading events at early stages using procedures that produce detailed and rapid genetic characterization of isolates.
Using the procedures described here, genetic and phylogenetic data was produced in response to an alert from syndrome surveillance within two to three weeks of receiving the alert. The presence of a large number of G12s reflects a global emergence of G12 [20,24]. The prevalence of G12 will need to be followed over the next few seasons to see whether its predominance was due to annual variations or to a partial escape from vaccine protection. Conducting different surveillance strategies in parallel (in this case syndrome surveillance, laboratory surveillance, and rapid genetic characterization of isolates) strengthens the reliability of interpreting data from any one of the platforms especially when sample sizes are small and strengthens our ability to detect and characterize potential spreading events at early stages.
We would like to thank Dani Cohen (TAU) and Tami Shohat (TAU, ICDC) for their helpful discussions. We especially acknowledge the contributions of Roberto Azar and Ilana Silberstein from the Israel National Center for Viral Gastroenteritis.
- Parashar UD, Gibson CJ, Bresee JS, Glass RI (2006) Rotavirus and severe childhood diarrhea. Emerg Infect Dis 12: 304-306. [Crossref]
- Parashar UD, Burton A, Lanata C, Boschi-Pinto C, Shibuya K, et al. (2009) Global mortality associated with rotavirus disease among children in 2004. J Infect Dis 200: S9-S15. [Crossref]
- Estes MK, Kapikian AZ (2007) Rotaviruses. (5th edn), Lippincott Williams & Wilkins, Philadelphia, pp: 1917-1974.
- Estes MK, Cohen J (1989) Rotavirus gene structure and function. Microbiol Rev 53: 410-449. [Crossref]
- Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, et al. (2006) Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 354: 11-22. [Crossref]
- Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, et al. (2006) Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 354: 23-33. [Crossref]
- Muhsen K, Shulman L, Kasem E, Rubinstein U, Shachter J, et al. (2010) Effectiveness of rotavirus vaccines for prevention of rotavirus gastroenteritis-associated hospitalizations in Israel: A case-control study. Hum Vaccin 6: 450-454. [Crossref]
- Muhsen K, Shulman L, Rubinstein U, Kasem E, Kremer A, et al. (2009) Incidence, characteristics, and economic burden of rotavirus gastroenteritis associated with hospitalization of israeli children <5 years of age, 2007-2008. J Infect Dis 200: S254-S263. [Crossref]
- Muhsen K, Anis E, Rubinstein U, Kassem E, Goren S, et al. (2017) Effectiveness of rotavirus pentavalent vaccine under a universal immunization program in Israel, 2011-2015: a case-control study. Clin Microbiol Infect 24: 53-59. [Crossref]
- Muhsen K, Kassem E, Rubenstein U, Goren S, Ephros M, et al. (2016) Incidence of rotavirus gastroenteritis hospitalizations and genotypes, before and five years after introducing universal immunization in Israel. Vaccine 34: 5916-5922. [Crossref]
- Muhsen K, Rubenstein U, Kassem E, Goren S, Schachter Y, et al. (2015) A significant and consistent reduction in rotavirus gastroenteritis hospitalization of children under five years of age, following the introduction of universal rotavirus immunization in Israel. Hum Vaccin Immunother 11: 2475-2482. [Crossref]
- Muhsen K, Kassem E, Rubinstein U, Schachter Y, Kremer A, et al. (2013) Incidence and characteristics of sporadic norovirus gastroenteritis associated with hospitalization of children less than 5 years of age in Israel. Pediatr Infect Dis J 32: 688-690. [Crossref]
- Shulman LM, Silberstein I, Alfandari J, Mendelson E (2011) Genotyping rotavirus RNA from archived rotavirus-positive rapid test strips. Emerg Infect Dis 17: 44-48. [Crossref]
- Gunasena S, Nakagomi O, Isegawa Y, Kaga E, Nakagomi T, et al. (1993) Relative frequency of VP4 gene alleles among human rotaviruses recovered over a 10-year period (1982-1991) from Japanese children with diarrhea. J Clin Microbiol 31: 2195-2197. [Crossref]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, et al. (1990) Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol 28: 276-282. [Crossref]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410. [Crossref]
- Maes P, Matthijnssens J, Rahman M, Van Ranst M (2009) RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9: 238. [Crossref]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882. [Crossref]
- Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, et al. (2010) Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 27: 2431-2436. [Crossref]
- Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, et al. (2007) Evolutionary history and global spread of the emerging g12 human rotaviruses. J Virol 81: 2382-2390. [Crossref]
- Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, et al. (2009) Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science 324: 1444-1447. [Crossref]
- Shulman LM, Manor Y, Sofer D, Mendelson E (2012) Bioterrorism and Surveillance for Infectious Diseases - Lessons from Poliovirus and Enteric Virus Surveillance. J Bioterr Biodef S4: 004.
- Dennehy PH (2008) Rotavirus vaccines: an overview. Clin Microbiol Rev 21: 198-208. [Crossref]
- Samajdar S, Varghese V, Barman P, Ghosh S, Mitra U, et al. (2006) Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J Clin Virol 36: 183-188. [Crossref]



