Analysis of 545 NIPT high-risk cases with high risk for trisomy 21 (T21), 18 (T18), and 13 (T13), as well as monosomy X (MX) from routine NIPT testing in a single prenatal center in Germany. Analysis was performed using the VeriSeq NIPT Solution v2 (Illumina Inc., USA). The assessment of true vs false positive results were based on clinical outcome data.
The average fetal fraction of 9.7% was within the expected range in T21 and MX but lower in T18 and T13. For all high-risk groups sensitivity and specificity was far above 99%. The positive predictive value (PPV) was highest at trisomy 21 with 94.1%, followed by trisomy 18 with 80.9%. For trisomy 13 and Monosomy X, the PPV was clearly lower at 60.5% and 65.6%, respectively. PPV was dependent on different indications and maternal age.
We could show that statistical tools of the method like the log likelihood ratio (LLR) score and T-Statistics value are important to distinguish between (clinical) false positive and true positive NIPT results in trisomies. The relationship between results and quality scores is less significant for MX cases.
The study shows that the Illumina VeriSeq v2 procedure is a highly reliable NIPT method with a low no call rate in the hands of experienced diagnostic laboratories.
fetal chromosomal aneuploidies, trisomy 21, trisomy 18, trisomy 13, monosomy X, fetal fraction, non-invasive prenatal testing (NIPT), positive predictive value (ppv), VeriSeq NIPT Solution
Noninvasive Prenatal Testing (NIPT) is a molecular genetic test that can determine whether a pregnancy is at high-risk for the common aneuploidies (trisomy 21, trisomy 18, trisomy 13, and sex chromosome aneuploidies) by analyzing circulating cell-free fetal DNA (cfDNA) present in maternal plasma. The first use of cfDNA for analysis of fetal genetic status, fetal sex determination based on the presence or absence of the Y chromosome, was reported in 1997 by Lo, et al. [1]. At that time, sequencing methods suitable for routine evaluation of millions of DNA fragments were not readily available nor was the corresponding analysis and evaluation software. Thus, cfDNA was not incorporated into routine clinical use at that time. Since that time there have been significant advantages in next-generation sequencing technology as well as the bioinformatics needed for analysis of sequencing results. These changes and commercialization of the cfDNA technology enabled the implementation of NIPT in a clinical setting close to a decade ago. This method initiated a revolution in prenatal testing: a move away from invasive options, such as amniocentesis and chorionic villus sampling (CVS), towards noninvasive prenatal screening.
In the 10 years since NIPT became available, it has become well accepted in developed regions [2]. Numerous studies have been published on the performance and use of NIPT. Most of these studies are designed as multicenter studies and include comparatively few abnormal trisomic pregnancies, with most samples originating from unaffected pregnancies. Here, we describe the clinical performance of NIPT within our single prenatal unit, which is the primary NIPT laboratory in Germany. The present work includes one of the largest NIPT evaluations of trisomies 21, 18, and 13 as well as monosomy X from a single prenatal unit and is intended to provide guidance in the evaluation of such results for genetic counselors, gynecologists, and diagnosticians in the laboratory.
Study patient/Sample details
Our full cohort included more than 40,000 singleton and twin pregnancies from a general German and Austrian pregnancy population that elected to undergo NIPT between December 2017 and February 2021; a subset of these cases were reported previously [3]. Within the full cohort, there were 545 cases reported by our NIPT assay at high risk for trisomies 21, 18, and 13, as well as monosomy X, henceforth referred to as the study cohort. Samples of at least 10 weeks of gestation were included in the study; exclusion criteria were a known vanishing twin or a higher-order multiple pregnancy. The amedes lab complies with the provisions of the German Federal Data Protection Act. All data were de-identified prior to enrollment in the study. Informed consent was obtained from all patients participating in the study to use their data for appropriate quality control and improvement of the NIPT assays.
Consistent with our previous study [3], indications for NIPT included advanced maternal age, a positive screening test result (ultrasound, serum markers), other medical reasons, or patient anxiety. Other medical reasons included abnormal ultrasound, a history of pregnancy complications including miscarriage or a previously affected pregnancy (e.g., trisomy 21, 18, 13, monosomy X), a genetic aberration in the family (e.g., trisomy 21), known diseases such as diabetes, epilepsy, and carcinoma, medications such as chemotherapy, or consanguinity. In the absence of information on the indication, patient anxiety or advanced maternal age (if the patient was ≥ 35 years) were used as indications. If multiple indications were provided, cases were assigned to a sole indication with the following priority: (1) positive screening test result, (2) advanced maternal age, (3) other medical reasons, (4) patient anxiety. The correctness of the assignment of cases to the different indication groups was dependent on the accuracy of the information received from the attending gynecologists/pediatricians, as was all other patient history information and feedback on the clinical pregnancy outcome.
Clinical results for the 545 high-risk study cohort cases were verified by invasive prenatal diagnostic procedures (cytogenetic analysis after CVS and/or amniocentesis), cytogenetic analysis of products of conception (POC) or placenta, postmortem examinations such as autopsy or macroscopic assessment of the abortion, postnatal cytogenetic analyses, as well as ultrasound and newborn examination. NIPT results that were positive for fetal aneuploidy were considered confirmed if they were validated by either invasive prenatal diagnostics or an abnormality observed on ultrasound that was consistent with the high-risk NIPT. Low-risk NIPT results were considered confirmed if the attending physician reported the birth of a healthy newborn lacking physical features or phenotypes associated with trisomy 21, 18, 13, or monosomy X.
VeriSeq NIPT solution v2 assay
The Illumina NIPT assay, VeriSeq NIPT Solution v2 (Illumina Inc., San Diego, CA, USA), was used for this study. This assay uses a paired-end sequencing technique and reports fetal sex, risk for trisomy 21 (T21), 18 (T18), and 13 (T13), as well as sex chromosome aneuploidies. We used a customized result reporting to meet amedes requirements: reporting of sex chromosomes was explicitly limited to monosomy X (MX) and XX/XY. Other sex chromosome aberrations are not analyzed for medical and ethical reasons. This NIPT test is offered by the amedes lab group in Germany and Austria under the name “fetalis”.
Data analysis was as previously described [3]. Briefly, a fetal fraction (FF) estimate was reported for each sample. Following data analysis, a log likelihood ratio (LLR) score was provided for each sample. This is the probability of a sample being affected given the sample’s estimated FF and observed coverage. The assay software also uses a dynamic threshold metric known as the individualized Fetal Aneuploidy Confidence Test (iFACT), which determines whether there is sufficient sequencing coverage for each individual sample given the FF estimate for that sample; samples that did not meet this threshold were reported as QC failures. A T-Statistics value is provided by the software and was used to help differentiate between low-risk and high-risk samples [3].
Statistics
Statistical data analysis was performed using Microsoft Excel 2016. Where applicable, statistical significance was assessed using a Student's t-test; a p value <0.05 was considered significant. Binomial 95% confidence intervals (CI) were calculated for sensitivity and specificity estimates.
In case of an inconclusive NIPT result, the analysis was repeated on the initial sample and/or on a second blood sample. In these cases, the result of the first analysis was excluded from the statistical analysis, so that only one result per patient was included in the statistical analysis. In addition, in patients where a NIPT result was obtained from the first blood sample but a repeat analysis was requested (e.g., due to a FF <2%), another blood sample was analyzed as a goodwill gesture. In these cases, the result of the first analysis was excluded from the statistical evaluation, except in cases where the repeat analysis did not yield a NIPT result; in the latter cases, the result of the first analysis was included in the statistics and the unsuccessful repeat analysis was excluded.
Demographics
Demographics of all patients analyzed are shown in Table 1. The 545 high-risk NIPT cases analyzed in the present study were part of a collection of 41,607 pregnancy cases, including 13,607 cases published previously [3]. The 41,607 cases included 40,871 (98.2%) singleton pregnancies and 736 (1.8%) twin pregnancies (Table 1). The mean maternal age was 33.5 years and the mean gestational age was 12.4 weeks. The primary indications for NIPT were patient anxiety (51%) and advanced maternal age (40%).
Table 1. Patient demographics for the full cohort
Study cohort |
Cases, n |
41,607 |
Singleton pregnancies, n (%) |
40,871 (98.2) |
Twin pregnancies, n (%) |
736 (1.8) |
Maternal age (year) |
Mean ± SEM |
33.45 ± 0.02 |
Range |
16.25 – 55.87 |
Gestational age (week) |
Mean ± SEM |
12.38 ± 0.01 |
Range |
10.00 – 36.57 |
BMI |
Mean ± SEM |
25.10 ± 0.03 |
Range |
14.60 – 62.44 |
Indication for screening |
Advanced maternal age, n (%) |
16,675 (40.1) |
Positive screening test result*, n (%) |
1,890 (4.5) |
Other medical reasons†, n (%) |
1,918 (4.6) |
Patient anxiety, n (%) |
21,124 (50.8) |
SEM: standard error of the mean; BMI: body mass index; *Positive screening test result includes ultrasound or serum marker screening; †Other medical reasons include e.g. abnormal ultrasound, known diseases of the patient (e.g., diabetes, epilepsy, and carcinoma), medication (e.g., chemotherapy), or a high-risk family medical history such as a previous miscarriage, a genetic aberration in a previous pregnancy (e.g., trisomy 21, 18, 13, monosomy X), a genetic aberration in the family (e.g., trisomy 21), or consanguinity.
Assay performance and clinical outcomes
Within 41,607 cases, 41,070 (98.7%) had an NIPT result with the first blood sample. The first pass failure rate was improved from 2.0% (821/ 41,607) to 1.3% (537/ 41,607) by repeated analysis of the initial sample. For the 537 pregnant women who did not get a result with the first blood draw, a repeated blood draw was offered, of which 459 (85.5%) patients accepted. Of these, 383 (83.4%) received an NIPT result with the second blood sample. Overall, a total of 41,453 (99.6%) patients obtained an NIPT result. The final failure rate was 0.4% (154/ 41,607). The mean BMI was significantly higher in cases without an NIPT result (32.79 ± 0.67) compared with the low-risk group (25.07 ± 0.03; p<0.001).
Of the 41,453 patients who received an NIPT result, 40,908 (98.3%) had a low-risk result and 545 (1.3%) patients were reported as at high-risk for a fetal chromosome aneuploidy (Table 2). Clinical outcomes were available for 7,012 (16.9%) cases; outcomes were based on invasive diagnostic techniques, ultrasound, or newborn physical exam. Based on clinical follow up for low-risk NIPT results (6,559/ 40,908), the negative predictive value (NPV) was > 99.9% (6,558/ 6,559). In one case, a false negative trisomy 21 result was observed, the fetal fraction of this false-negative case was 3%.
Table 2. Cases of clinical follow up, negative predictive value (NPV) for low-risk NIPT results and sensitivity, specificity, and positive predictive value (PPV) for total high-risk NIPT cases
Total cases, n |
41,607 |
Low-risk |
Cases, n (%) |
40,908 (98.3) |
Cases with follow-up (N), n (%) |
6,559 (16.0) |
TN, n (%) |
6,558 (99.98) |
FN, n (%) |
1 (0.02) |
NPV, % (TN/TP+FN) |
>99.9 (6,558/ 6,559) |
High-risk |
Cases, n (%) |
545 (1.3) |
Cases with follow-up (N), n (%) |
452 (82.9) |
Sensitivity, % (95% CI)
(TP/TP+FN) |
99.7 (98.6 – 100.0)
(391/392) |
Specificity, % (95% CI)
(TN+TP/TN+TP+FP) |
99.1 (98.9 – 99.3)
(6,949/ 7,010) |
PPV, % (TP/N) |
86.5 (391/ 452) |
Theoretical lower PPV, %
(TP/TP+FP+unk) |
76.1
(391/ 514) |
Theoretical upper PPV, %
(TP+unk/TP+FP+unk) |
88.1
(453/ 514) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
The 545 cases with a high-risk NIPT result were actively followed up for clinical outcome information. In 82.9% cases (452/ 545), outcome information was obtained. There were 93 cases classified as having unknown clinical outcomes: In 62 cases, clinical outcome information could not be obtained, or the pregnancy was terminated or ended in miscarriage, but no information on the fetal genetic constellation was available; 27 patients were recent cases and follow up diagnostic testing has not yet been performed; four patients decided to continue their pregnancies and no further information is available. Based on the high-risk NIPT results cases with known outcomes, this NIPT has high overall sensitivity (99.7%), specificity (99.1%), and positive predictive value (PPV; 86.5%) (Table 2).
Of the 545 high-risk NIPT results, trisomy 21 was the most commonly reported aneuploidy (61.5%), followed by trisomy 18 (20.6%), trisomy 13 (9.7%), and monosomy X (8.1%); there was one case reported as a double aneuploidy (Table 3). Sensitivity and specificity were far above 99% for each aneuploidy. The PPV was highest for trisomy 21 at 94.1%, followed by trisomy 18 at 80.9%. The PPVs were lower for trisomy 13 (60.5%) and monosomy X (65.6%) (Table 3).
Table 3. Sensitivities, specificities, and positive predictive values (PPV) for high-risk NIPT cases
High-risk NIPT result |
Trisomy 21 |
Trisomy 18 |
Trisomy 13 |
Trisomy 13 + 21 |
Monosomy X |
Cases, n (%) |
335 (61.5) |
112 (20.6) |
53 (9.7) |
1 (0.2) |
44 (8.1) |
Cases with follow-up (N), n (%) |
287 (85.7) |
94 (83.9) |
38 (71.7) |
1 (-) |
32 (72.7) |
Sensitivity, % (95% CI)
(TP/TP+FN) |
99.6 (98.0 – 100.0)
(270/ 271) |
>99,9 (95.3 – 100.0)
(76/ 76) |
>99,9 (85.2 – 100.0)
(23/ 23) |
-
(1/ 1) |
>99.9 (83.9 – 100.0)
(21/21) |
Specificity, % (95% CI)
(TN+TP/TN+TP+FP) |
99.8 (99.6 – 99.9)
(6,828/ 6,845) |
99.7 (99.6 – 99.8)
(6,634/ 6,652) |
99.8 (99.6 – 99.9)
(6,581/ 6,596) |
-
(6,559/ 6,559) |
99.8 (99.7 – 99.9)
(6,579/ 6,590) |
PPV, % (TP/N) |
94.1 (270/ 287) |
80.9 (76/ 94) |
60.5 (23/ 38) |
- (1/ 1) |
65.6 (21/ 32) |
Theoretical lower PPV, %
(TP/TP+FP+unk) |
85.2
(270/ 317) |
70.4
(76/ 108) |
44.2
(23/ 52) |
-
(1/ 1) |
58.3
(21/ 36) |
Theoretical upper PPV, %
(TP+unk/TP+FP+unk) |
94.6
(300/ 317) |
83.3
(90/ 108) |
71.2
(37/ 52) |
-
(1/ 1) |
69.4
(25/ 36) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Analysis of the high-risk NIPT results with respect to indication was to investigate differences between high- and low-risk populations. This analysis showed that specificity and sensitivity levels were >99.0% regardless of indication (Table 4). For each aneuploidy category, the PPV in the advanced maternal age, high-risk screening, and other medical reasons indication groups were higher than for the anxiety indication. In trisomy 18, trisomy 13, and monosomy X high-risk NIPT results, the PPV in the high-risk screening and other medical reasons indication groups were above the PPV of the advanced maternal age group. For trisomy 21 cases, the highest PPVs were observed in the high-risk screening (98.6%) and advanced maternal age (95.0%) groups.
Table 4. Sensitivities, specificities and PPV values for high-risk cases stratified by indication
Indication |
Adv. maternal age |
Screening |
Other med. reasons |
Anxiety |
Cases, n (%) |
16,675 (40.1) |
1,890 (4.5) |
1,918 (4.6) |
21,124 (50.8) |
Total high-risk cases, n (%) |
287 (1.7) |
125 (6.6) |
23 (1.2) |
110 (0.5) |
Trisomy 21 |
Cases, n (%) |
185 (55.2) |
83 (24.8) |
13 (3.9) |
54 (16.1) |
Cases with follow-up (N), n (%) |
161 (87.0) |
72 (86.7) |
9 (69.2) |
45 (83.3) |
Sensitivity, % (TP/TP+FN) |
>99.9 (153/ 153) |
>99.9 (71/ 71) |
>99.9 (8/ 8) |
97.4 (38/ 39) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,698/ 2,706) |
99.6 (256/ 257) |
99.6 (282/ 283) |
99.8 (3,592/ 3,599) |
PPV, % (TP/N) |
95.0 (153/ 161) |
98.6 (71/ 72) |
88.9 (8/ 9) |
84.4 (38/ 45) |
Trisomy 18 |
Cases, n (%) |
62 (55.4) |
25 (22.3) |
4 (3.6) |
21 (18.8) |
Cases with follow-up (N), n (%) |
57 (91.9) |
20 (80.0) |
3 (75.0) |
14 (66.7) |
Sensitivity, % (TP/TP+FN) |
>99.9 (49/ 49) |
>99.9 (18/ 18) |
>99.9 (3/ 3) |
>99.9 (6/ 6) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,594/ 2,602) |
99.0 (203/ 205) |
>99.9 (277/ 377) |
99.8 (3,560/ 3,568) |
PPV, % (TP/N) |
86.0 (49/ 57) |
90.0 (18/ 20) |
>99.9 (3/ 3) |
42.9 (6/ 14) |
Trisomy 13 |
Cases, n (%) |
26 (49.1) |
8 (15.1) |
2 (3.8) |
17 (32.1) |
Cases with follow-up (N), n (%) |
17 (65.4) |
6 (75.0) |
2 (100.0) |
13 (76.5) |
Sensitivity, % (TP/TP+FN) |
>99.9 (10/ 10) |
>99.9 (4/ 4) |
>99.9 (2/ 2) |
>99.9 (7/ 7) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,555/ 2,562) |
99.0 (189/ 191) |
>99.9 (276/ 276) |
99.8 (3,561/ 3,567) |
PPV, % (TP/N) |
58.8 (10/ 17) |
66.7 (4/ 6) |
>99.9 (2/ 2) |
53.8 (7/ 13) |
Monosomy X |
Cases, n (%) |
13 (29.5) |
9 (20.5) |
4 (9.1) |
18 (40.9) |
Cases with follow-up (N), n (%) |
9 (69.2) |
7 (77.8) |
3 (75.0) |
13 (72.2) |
Sensitivity, % (TP/TP+FN) |
>99.9 (5/ 5) |
>99.9 (7/ 7) |
>99.9 (3/3) |
>99.9 (6/ 6) |
Specificity, % (TN+TP/TN+TP+FP) |
99.8 (2,550/ 2,554) |
>99.9 (192/ 192) |
>99.9 (277/277) |
99.8 (3,560/ 3,567) |
PPV, % (TP/N) |
55.6 (5/ 9) |
>99.9 (7/ 7) |
>99.9 (3/3) |
46.2 (6/ 13) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
To investigate the role of maternal age in high-risk NIPT results, the relationship between different maternal age groups and sensitivity, specificity, and PPV was evaluated (Table 5). The sensitivity was ≥ 98.6% and specificity was well above 99.0% in each age group. For cases at high risk for trisomy 21, 18, and 13, the PPV was higher in cases with an advanced maternal age (≥ 35 years) compared with younger women (<35 years). This was particularly true for cases reported as trisomy 21, where PPVs increased from 78.9% (25 to 29 years) and 93.0% (30 to 34 years) to 96.7% (35 to 39 years) and 94.3% (>40 years). In the monosomy X risk group, an association with maternal age was not evident.
Table 5. Sensitivities, specificities and PPV values for high-risk cases stratified by maternal age
Maternal age, years |
< 25 |
25 – 29 |
30 – 34 |
35 – 39 |
> 40 |
Cases, n (%) |
1,057 (2.5) |
6,889 (16.6) |
16,099 (38.7) |
14,020 (33.7) |
3.541 (8.5) |
Total high-risk cases, n (%) |
8 (0.0) |
41 (0.1) |
125 (0.3) |
220 (0.5) |
151 (0.4) |
Trisomy 21, 18, 13, 13 + 21 |
Cases, n (%) |
6 (1.2) |
36 (7.2) |
103 (20.6) |
207 (41.3) |
149 (29.7) |
Cases with follow-up (N), n (%) |
5 (83.3) |
29 (80.6) |
83 (80.6) |
172 (83.1) |
131 (87.9) |
Sensitivity, % (TP/TP+FN) |
>99.9 (2/ 2) |
>99.9 (22/ 22) |
98.6 (71/ 72) |
>99.9 (160/ 160) |
>99.9 (115/ 115) |
Specificity, % (TN+TP/TN+TP+FP) |
98.4 (180/ 183) |
99.4 (1,190/ 1,197) |
99.5 (2,653/ 2,665) |
99.5 (2,315/ 2,327) |
97.4 (590/ 606) |
PPV, % (TP/N) |
40.0 (2/ 5) |
75.9 (22/ 29) |
85.5 (71/ 83) |
93.0 (160/ 172) |
87.8 (115/ 131) |
Monosomy X |
Cases, n (%) |
2 (0.6) |
5 (1.5) |
22 (6.6) |
13 (3.9) |
2 (0.6) |
Cases with follow-up (N), n (%) |
1 (50.0) |
3 (60.0) |
17 (77.3) |
9 (69.2) |
2 (100.0) |
Sensitivity, % (TP/TP+FN) |
- (1/ 1) |
>99.9 (2/ 2) |
>99.9 (11/ 11) |
>99.9 (5/ 5) |
>99.9 (2/ 2) |
Specificity, % (TN+TP/TN+TP+FP) |
- (179/ 179) |
99.9 (1,170/ 1,171) |
99.8 (2,593/ 2,599) |
99.8 (2,160/ 2,164) |
>99.9 (477/ 477) |
PPV, % (TP/N) |
- (1/ 1) |
66.7 (2/ 3) |
64.7 (11/ 17) |
55.6 (5/ 9) |
>99.9 (2/ 2) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Analysis of the incidence of the high-risk results for trisomies 21, 18, and 13 as well as monosomy X showed no significant association with gestational age at the time of sample collection for NIPT (Table 6).
Table 6. Sensitivities, specificities and PPV values for high-risk cases stratified by gestational age
Gestational age, week+day |
10+0 – 10+6 |
11+0 – 13+6 |
14+0 – 19+6 |
>20+0 |
Cases, n (%) |
7,556 (18.2) |
29,212 (70.2) |
4,392 (10.6) |
447 (1.1) |
Total high-risk cases, n (%) |
104 (1.4) |
371 (4.9) |
48 (0.6) |
22 (0.3) |
Trisomy 21, 18, 13, 13 + 21 |
Cases, n (%) |
93 (18.6) |
345 (68.9) |
44 (8.8) |
19 (3.8) |
Cases with follow-up (N), n (%) |
74 (79.6) |
291 (84.3) |
40 (90.9) |
15 (78.9) |
Sensitivity, % (TP/TP+FN) |
>99.9 (68/ 68) |
99.6 (257/ 258) |
>99.9 (30/ 30) |
>99.9 (15/ 15) |
Specificity, % (TN+TP/TN+TP+FP) |
99.5 (1,248/ 1,254) |
99.3 (4,922/ 4,956) |
98.6 (691/ 701) |
>99.9 (67/ 67) |
PPV, % (TP/N) |
91.9 (68/ 74) |
88.3 (257/ 291) |
75.0 (30/ 40) |
>99.9 (15/ 15) |
Monosomy X |
Cases, n (%) |
11 (25.0) |
26 (59.1) |
4 (9.1) |
3 (6.8) |
Cases with follow-up (N), n (%) |
9 (81.8) |
17 (65.4) |
4 (100.0) |
2 (66.7) |
Sensitivity, % (TP/TP+FN) |
>99.9 (6/ 6) |
>99.9 (12/ 12) |
>99.9 (2/ 2) |
- (1/ 1) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (1,186/ 1,189) |
99.9 (4,677/ 4,682) |
99.7 (663/ 665) |
- (53/ 54) |
PPV, % (TP/N) |
66.7 (6/ 9) |
70.6 (12/ 17) |
50.0 (2/ 4) |
- (1/ 2) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Fetal fraction
The average FF was 9.7% in low-risk pregnancies. The average FF was slightly lower in cases reported as trisomy 21 (9.4%), markedly lower for cases reported as trisomy 18 (7.2%) and trisomy 13 (6.0%), and unchanged in cases reported as monosomy X (10.1%) (Figure 1). We also evaluated performance in three FF subgroups: <4%, 4%-8%, and > 8% (Table 7). For cases reported as trisomy 21, the FF was above 4% in around 95% of cases and above 8% in about half the cases; only a 5.7% had an FF less than 4%. The specificity and sensitivity for trisomy 21 was >99% when the FF was at least 4%. The PPV increased with increasing FF from 71% to 99%. For trisomy 18 and trisomy 13, most cases had a FF ≤ 8%. Importantly, sensitivity and specificity levels were very high regardless of FF, >99.9% and ≥ 98.3%, respectively. Similar to trisomy 21, the PPV for trisomy 18 increased with increasing FF. The same association between PPV and FF was not observed for trisomy 13. All monosomy X cases had at least 4% FF, and all specificity and sensitivity levels were very high regardless of FF.
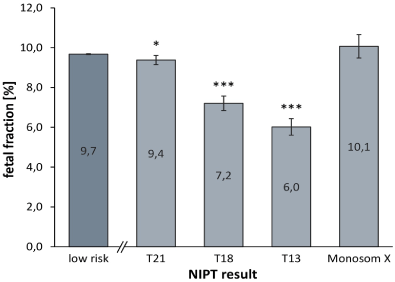
Figure 1. Chromosomal risk status and fetal fraction
Table 7. Sensitivities, specificities and PPV values for high-risk cases for cases with fetal fractions < 4%, 4%-8%, and > 8%
Fetal fraction, % |
< 4% |
4% – 8% |
> 8% |
Cases, n (%) |
1,015 (2.4) |
17,522 (42.3) |
22,916 (55.3) |
Total high-risk cases, n (%) |
47 (4.6) |
266 (1.5) |
232 (1.0) |
Trisomy 21 |
Cases, n (%) |
19 (5.7) |
148 (44.2) |
168 (50.1) |
Cases with follow-up (N), n (%) |
14 (73.7) |
127 (85.8) |
146 (86.9) |
Sensitivity, % (TP/TP+FN) |
90.9 (10/ 11) |
>99.9 (116/ 116) |
>99.9 (144/ 144) |
Specificity, % (TN+TP/TN+TP+FP) |
97.8 (180/ 184) |
99.6 (3,094/ 3,105) |
99.9 (3,554/ 3,556) |
PPV, % (TP/N) |
71.4 (10/ 14) |
91.3 (116/ 127) |
98.6 (144/ 146) |
Trisomy 18 |
Cases, n (%) |
15 (13.4) |
64 (57.1) |
33 (29.5) |
Cases with follow-up (N), n (%) |
10 (66.7) |
57 (89.1) |
27 (81.8) |
Sensitivity, % (TP/TP+FN) |
>99.9 (7/ 7) |
>99.9 (45/ 45) |
>99.9 (24/ 24) |
Specificity, % (TN+TP/TN+TP+FP) |
98.3 (177/ 180) |
99.6 (3,023/ 3,035) |
99.9 (3,434/ 3,437) |
PPV, % (TP/N) |
70.0 (7/ 10) |
78.9 (45/ 57) |
88.9 (24/ 27) |
Trisomy 13 |
Cases, n (%) |
13 (24.5) |
32 (60.4) |
8 (15.1) |
Cases with follow-up (N), n (%) |
9 (69.2) |
23 (71.9) |
6 (75.0) |
Sensitivity, % (TP/TP+FN) |
>99.9 (6/ 6) |
>99.9 (14/ 14) |
>99.9 (3/ 3) |
Specificity, % (TN+TP/TN+TP+FP) |
98.3 (176/ 179) |
99.7 (2,992/ 3,001) |
99.9 (3,413/ 3,416) |
PPV, % (TP/N) |
66.7 (6/ 9) |
60.9 (14/ 23) |
50.0 (3/ 6) |
Monosomy X |
Cases, n (%) |
0 (0.0) |
22 (50.0) |
22 (50.0) |
Cases with follow-up (N), n (%) |
- |
17 (77.3) |
15 (68.2) |
Sensitivity, % (TP/TP+FN) |
- (0/ 0) |
>99.9 (12/ 12) |
>99.9 (9/ 9) |
Specificity, % (TN+TP/TN+TP+FP) |
- (170/ 170) |
99.8 (2,990/ 2,995) |
99.8 (3,419/ 3,425) |
PPV, % (TP/N) |
- (0/ 0) |
70.6 (12/ 17) |
60.0 (9/ 15) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
By analyzing the LLR score, which is a degree of probability that a fetal aberration is present, it became obvious that all true positive trisomy high-risk cases had approximately 3-8 fold higher LLR scores and 1.8–3.4 fold higher T-Statistics values compared to false positive cases (Table 8). No differences were found between true positive and false positive monosomy X cases.
Table 8. Relationship between NIPT results and LLR scores/T-Statistics values
|
True positive |
False positive |
|
LLR Score |
T-Statistics value |
LLR Score |
T-Statistics value |
Trisomy 21 |
179.1 ± 9.7 |
16.6 ± 0.5 |
20.9 ± 9.8 |
4.9 ± 0.9 |
Trisomy 18 |
170.5 ± 19.3 |
16.2 ± 1.0 |
44.1 ± 18.0 |
8.0 ± 1.6 |
Trisomy 13 |
126.6 ± 21.7 |
14.6 ± 1.5 |
47.1 ± 17.2 |
8.6 ± 1.6 |
Monosomy X |
50.8 ± 8.5 |
-24.2 ± 2.2 |
48.8 ± 18.9 |
-22.0 ± 4.4 |
Values shown as mean ± standard error of the mean (SEM).
NIPT for trisomy 21, 18, and 13 as well as fetal sex and sex chromosome aberrations has been rapidly adopted into clinical practice worldwide. NIPT in Germany, Austria, and Switzerland is regulated by a number of professional societies, including the German Society of Human Genetics [4,5], the German Society for Ultrasound in Medicine (DEGUM), Austrian Society for Ultrasound in Medicine (ÖGUM), Swiss Society for Ultrasound in Medicine (SGUM), and Fetal Medicine Foundation (FMF) Germany [6,7]. Our laboratory follows these guidelines and recommendations. Here we showed the low failure rate of 0.4% and high performance of the NIPT assay used in our laboratory across maternal ages, gestational ages, and FF values in a large clinical population.
NIPT is increasingly being adopted by smaller laboratories in their routine services. There is a need for larger collections of aneuploid cases within individual laboratories to provide sufficient experience to evaluate questionable findings and to serve as a knowledge base to enable a laboratory to advise the referring gynecologists and the patients about the significance of the NIPT findings. The amedes genetic unit is one of the largest labs in Germany. We have been performing genetic counselling, amniocentesis and CVS for over 30 years, first trimester screening for 20 years, and we have 10 years experience with NIPT. During our time performing NIPT, we have used each of the major NIPT methods in our lab and have chosen to use the Illumina VeriSeq NIPT Solution platform for the last 3 years. With this platform, we have processed more than 40,000 samples, including 545 reported as high-risk for a fetal chromosome aneuploidy. Follow up was obtained for 82.9% of the high-risk cases. Therefore, we feel the high performance values determined here are a reliable estimate of the true performance of this assay in a clinical setting. The likelihood that the PPV in particular could shift in one direction or the other is relatively low, but it is possible that some subgroups are not a reliable estimate given the low number of cases. Since the sensitivities and specificities were well above 99% for all high-risk aneuploidy groups studied, this data supports the accuracy of NIPT in general as previously reported in a systematic review [8]. The technical data presented here must not be taken out of context of clinical findings. Irreversible clinical decisions should not be made based on NIPT results alone. High-risk NIPT results should always be confirmed by amniocentesis or CVS.
The Illumina evaluation software includes tools like the LLR score and the T-Statistics value. In our daily work, these parameters have proved to be very helpful in discriminating between true and false positive cases for the common trisomies. We use this valuable information in the consultation of each case.
We think our subgroup analyses for PPVs are of particular interest and value. Consistent with the literature [9,10], the highest PPV was for trisomy 21, with lower values for trisomy 18 and trisomy 13. This finding can be explained by the difference in prevalence between the trisomies: the highest in trisomy 21 and the lowest in trisomy 13. Previous studies [11,12] have shown that the PPV for monosomy X cases reported by NIPT are in the low [13] to medium range [14], as was also observed in this study. What is the reason for this? One study reported that 8.6% of positive results for sex chromosome aneuploidy were due to maternal mosaicism [15]. Such maternal sex chromosome abnormalities may cause discordant results between NIPT result and the fetus [16]. But an early detection of phenotype-genotype-sex discordance is important to find evidence of underlying genetic, chromosomal, or biochemical disease, and also to enable time-critical postnatal treatment [17]. They emphasized the need for simultaneous detailed ultrasound examination to detect and quickly clarify discrepant situations. Importantly, as the NIPT method used here incorporates fragment length information, this reduces the likelihood of maternal chromosome anomalies causing a high-risk call by NIPT.
If we look at the impact of different indications on the PPVs we can see that the higher the basic risk of an indication, the higher the PPV. For example, the PPVs for the indication "increased maternal age" or "high-risk screening" are significantly higher than for the indication "anxiety". This is likely explained by a lower background prevalence of fetal aneuploidy in the lower risk (anxiety) indication group [18]. Women from low-risk indications and younger women should be counseled about a lower PPV.
Fetal fraction is one of the most important quality control parameters for NIPT and should be considered for proper counseling and further clinical management [19]. The average FF is around 10%. The FF depends on factors, such as the maternal BMI (FF decreases with increasing BMI) [20,21], the type of trisomy (lower FF in trisomies 13 and 18) [22], and the presence of mosaicism [23]. Significantly lower FF was found also in this study in cases screened positive for trisomy 18 and trisomy 13. Different NIPT approaches have different minimum threshold values of FF for reporting, with some failing all samples below a set threshold (between 2% and 4%) [19]. For this reason, it is very important to choose an NIPT platform that includes effective FF quality metrics to minimize failures linked to technical reasons [24]. The method employed here uses a dynamic threshold metric named iFACT, which determines whether there is sufficient sequencing coverage for each individual sample given the FF estimate for that sample. This metric enables accurate reporting at low fetal fractions and a low failure rate. This is evident from the data reported here where fetal aneuploidies were accurately reported at FF below 4%. This is particularly important for trisomy 13 and 18 as the average FF is much lower for these aneuploidies. The overall no call rate for the initial blood sample in this study was 1.3% and use of a second blood sample reduced the failure rate to a low 0.4%. Thus, 99.6% of all 41,607 pregnant women received an NIPT result. To our knowledge this is the lowest no call rate reported.
Our experience with over 40,000 clinical NIPT cases shows that the present NGS-based NIPT method can reliably identify the most frequent aneuploidies occurring prenatally. Fortunately, the majority of NIPT results are normal, so that these results lead to a rapid relief of the pregnant women.
Wolf Kupatt and Christoph Keck (amedes) have always supported and advanced the NIPT project excellently. In addition, we want to thank the many gynaecologists who gave us valuable information on the fetal outcome.
Assistance with data analysis and paper preparation was provided by Illumina, Inc. but no financial renumeration was provided directly to the authors.
The authors declare that they have no conflict of interest.
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, et al. (1997) Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485-487. [Crossref]
- Gadsboll K, Petersen OB, Gatinois V, Strange H, Jacobsson B, et al. (2020) Current use of noninvasive prenatal testing in Europe, Australia and the USA: A graphical presentation. Acta Obstet Gynecol Scand 99: 722-730. [Crossref]
- Borth H, Teubert A, Glaubitz R, Knippenberg S, Kutur N, et al. (2020) Analysis of cell-free DNA in a consecutive series of 13,607 routine cases for the detection of fetal chromosomal aneuploidies in a single center in Germany. Arch Gynecol Obstet 2020.
- https://www.bvnp.de/article/8-konsenspapier-nipt-aufnahme-in-den-leistungskatalog-der-g-kv-thesen-der-dggg-der/
- https://www. gfhev.de/leitlinien
- Kozlowski P, Burkhardt T, Gembruch U, Gonser M, Kahler C, et al. (2019) DEGUM, OGUM, SGUM and FMF Germany recommendations for the implementation of first-trimester screening, detailed ultrasound, cell-free DNA screening and diagnostic procedures. Ultraschall Med 40: 176-193. [Crossref]
- Schmid M, Klaritsch P, Arzt W, Burkhardt T, Duba HC, et al. (2015) Cell-free DNA testing for fetal chromosomal anomalies in clinical practice: Austrian-German-Swiss recommendations for non-invasive prenatal tests (NIPT). Ultraschall Med 36: 507-510. [Crossref]
- Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH (2017) Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 50: 302-314. [Crossref]
- Taneja PA, Snyder HL, de Feo E, Kruglyak KM, Halks-Miller M, et al. (2016) Noninvasive prenatal testing in the general obstetric population: clinical performance and counseling considerations in over 85 000 cases. Prenat Diagn 36: 237-243. [Crossref]
- Xu L, Huang H, Lin N, Wang Y, He D, et al. (2020) Non-invasive cell-free fetal DNA testing for aneuploidy: multicenter study of 31 515 singleton pregnancies in southeastern China. Ultrasound Obstet Gynecol 55: 242-247. [Crossref]
- Zhang B, Lu BY, Yu B, Zheng FX, Zhou Q, et al. (2017) Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J Int Med Res 45: 621-630. [Crossref]
- Eiben B, Krapp M, Borth H, Kutur N, Kreiselmaier P, et al. (2015) Single Nucleotide Polymorphism-Based Analysis of Cell-Free Fetal DNA in 3000 Cases from Germany and Austria. Ultrasound Int Open 1: E8-E11. [Crossref]
- McLennan A, Palma-Dias R, da Silva Costa F, Meagher S, Nisbet DL, Scott F (2016) Noninvasive prenatal testing in routine clinical practice--an audit of NIPT and combined first-trimester screening in an unselected Australian population. Aust N Z J Obstet Gynaecol 56: 22-28. [Crossref]
- Kornman L, Palma-Dias R, Nisbet D, Scott F, Menezes M, et al. (2018). Non-invasive prenatal testing for sex chromosome aneuploidy in routine clinical practice. Fetal Diagn Ther 44: 85-90. [Crossref]
- Wang Y, Chen Y, Tian F, Zhang J, Song Z, et al. (2014) Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem 60: 251-259. [Crossref]
- Martin KA, Samango-Sprouse CA, Kantor V, Dhamankar R, Valenti E, et al. (2020) Detection of maternal X chromosome abnormalities using single nucleotide polymorphism-based noninvasive prenatal testing. Am J Obstet Gynecol MFM 2: 100152. [Crossref]
- Smet ME, Scott FP, McLennan AC (2020) Discordant fetal sex on NIPT and ultrasound. Prenat Diagn 40: 1353-1365. [Crossref]
- Hui L, Bianchi DW (2017) Noninvasive Prenatal DNA Testing: The Vanguard of Genomic Medicine. Annu Rev Med 68: 459-472. [Crossref]
- Hui L, Bianchi DW (2020) Fetal fraction and noninvasive prenatal testing: What clinicians need to know. Prenat Diagn 40: 155-163. [Crossref]
- Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH (2013) Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol 41: 26-32. [Crossref]
- Panchalee T, Vossaert L, Wang Q, Crovetti BR, McCombs AK, et al. (2020) The effect of maternal body mass index and gestational age on circulating trophoblast yield in cell-based noninvasive prenatal testing. Prenat Diagn 40: 1383-1389.
- Eiben B, Krapp M, Borth H, Kutur N, Kreiselmaier P, et al. (2015) Single Nucleotide Polymorphism-Based Analysis of Cell-Free Fetal DNA in 3000 Cases from Germany and Austria. Ultrasound Int Open 1: E8-E11. [Crossref]
- Brison N, Neofytou M, Dehaspe L, Bayindir B, Van Den Bogaert K, et al. (2018) Predicting fetoplacental chromosomal mosaicism during non-invasive prenatal testing. Prenat Diagn 38: 258-266. [Crossref]
- Grati FR, Kagan KO (2016) No test result rate of cfDNA analysis and its influence on test performance metrics. Ultrasound in Obstetrics and Gynecology 50: 134-137.
Editorial Information
Editor-in-Chief
Alessandro Buda
University of Milano Bicocca
Italy
Article Type
Research Article
Publication history
Received: February 25, 2021
Accepted: March 08, 2021
Published: March 11, 2021
Copyright
©2021 Eiben B. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Eiben B (2021) Clinical experience with noninvasive prenatal testing in Germany: Analysis of over 500 high-risk cases for trisomy 21, 18, 13 and monosomy X. Obstet Gnecol Rep 5(1): DOI: 10.15761/OGR.1000157.

Figure 1. Chromosomal risk status and fetal fraction
Table 1. Patient demographics for the full cohort
Study cohort |
Cases, n |
41,607 |
Singleton pregnancies, n (%) |
40,871 (98.2) |
Twin pregnancies, n (%) |
736 (1.8) |
Maternal age (year) |
Mean ± SEM |
33.45 ± 0.02 |
Range |
16.25 – 55.87 |
Gestational age (week) |
Mean ± SEM |
12.38 ± 0.01 |
Range |
10.00 – 36.57 |
BMI |
Mean ± SEM |
25.10 ± 0.03 |
Range |
14.60 – 62.44 |
Indication for screening |
Advanced maternal age, n (%) |
16,675 (40.1) |
Positive screening test result*, n (%) |
1,890 (4.5) |
Other medical reasons†, n (%) |
1,918 (4.6) |
Patient anxiety, n (%) |
21,124 (50.8) |
SEM: standard error of the mean; BMI: body mass index; *Positive screening test result includes ultrasound or serum marker screening; †Other medical reasons include e.g. abnormal ultrasound, known diseases of the patient (e.g., diabetes, epilepsy, and carcinoma), medication (e.g., chemotherapy), or a high-risk family medical history such as a previous miscarriage, a genetic aberration in a previous pregnancy (e.g., trisomy 21, 18, 13, monosomy X), a genetic aberration in the family (e.g., trisomy 21), or consanguinity.
Table 2. Cases of clinical follow up, negative predictive value (NPV) for low-risk NIPT results and sensitivity, specificity, and positive predictive value (PPV) for total high-risk NIPT cases
Total cases, n |
41,607 |
Low-risk |
Cases, n (%) |
40,908 (98.3) |
Cases with follow-up (N), n (%) |
6,559 (16.0) |
TN, n (%) |
6,558 (99.98) |
FN, n (%) |
1 (0.02) |
NPV, % (TN/TP+FN) |
>99.9 (6,558/ 6,559) |
High-risk |
Cases, n (%) |
545 (1.3) |
Cases with follow-up (N), n (%) |
452 (82.9) |
Sensitivity, % (95% CI)
(TP/TP+FN) |
99.7 (98.6 – 100.0)
(391/392) |
Specificity, % (95% CI)
(TN+TP/TN+TP+FP) |
99.1 (98.9 – 99.3)
(6,949/ 7,010) |
PPV, % (TP/N) |
86.5 (391/ 452) |
Theoretical lower PPV, %
(TP/TP+FP+unk) |
76.1
(391/ 514) |
Theoretical upper PPV, %
(TP+unk/TP+FP+unk) |
88.1
(453/ 514) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 3. Sensitivities, specificities, and positive predictive values (PPV) for high-risk NIPT cases
High-risk NIPT result |
Trisomy 21 |
Trisomy 18 |
Trisomy 13 |
Trisomy 13 + 21 |
Monosomy X |
Cases, n (%) |
335 (61.5) |
112 (20.6) |
53 (9.7) |
1 (0.2) |
44 (8.1) |
Cases with follow-up (N), n (%) |
287 (85.7) |
94 (83.9) |
38 (71.7) |
1 (-) |
32 (72.7) |
Sensitivity, % (95% CI)
(TP/TP+FN) |
99.6 (98.0 – 100.0)
(270/ 271) |
>99,9 (95.3 – 100.0)
(76/ 76) |
>99,9 (85.2 – 100.0)
(23/ 23) |
-
(1/ 1) |
>99.9 (83.9 – 100.0)
(21/21) |
Specificity, % (95% CI)
(TN+TP/TN+TP+FP) |
99.8 (99.6 – 99.9)
(6,828/ 6,845) |
99.7 (99.6 – 99.8)
(6,634/ 6,652) |
99.8 (99.6 – 99.9)
(6,581/ 6,596) |
-
(6,559/ 6,559) |
99.8 (99.7 – 99.9)
(6,579/ 6,590) |
PPV, % (TP/N) |
94.1 (270/ 287) |
80.9 (76/ 94) |
60.5 (23/ 38) |
- (1/ 1) |
65.6 (21/ 32) |
Theoretical lower PPV, %
(TP/TP+FP+unk) |
85.2
(270/ 317) |
70.4
(76/ 108) |
44.2
(23/ 52) |
-
(1/ 1) |
58.3
(21/ 36) |
Theoretical upper PPV, %
(TP+unk/TP+FP+unk) |
94.6
(300/ 317) |
83.3
(90/ 108) |
71.2
(37/ 52) |
-
(1/ 1) |
69.4
(25/ 36) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 4. Sensitivities, specificities and PPV values for high-risk cases stratified by indication
Indication |
Adv. maternal age |
Screening |
Other med. reasons |
Anxiety |
Cases, n (%) |
16,675 (40.1) |
1,890 (4.5) |
1,918 (4.6) |
21,124 (50.8) |
Total high-risk cases, n (%) |
287 (1.7) |
125 (6.6) |
23 (1.2) |
110 (0.5) |
Trisomy 21 |
Cases, n (%) |
185 (55.2) |
83 (24.8) |
13 (3.9) |
54 (16.1) |
Cases with follow-up (N), n (%) |
161 (87.0) |
72 (86.7) |
9 (69.2) |
45 (83.3) |
Sensitivity, % (TP/TP+FN) |
>99.9 (153/ 153) |
>99.9 (71/ 71) |
>99.9 (8/ 8) |
97.4 (38/ 39) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,698/ 2,706) |
99.6 (256/ 257) |
99.6 (282/ 283) |
99.8 (3,592/ 3,599) |
PPV, % (TP/N) |
95.0 (153/ 161) |
98.6 (71/ 72) |
88.9 (8/ 9) |
84.4 (38/ 45) |
Trisomy 18 |
Cases, n (%) |
62 (55.4) |
25 (22.3) |
4 (3.6) |
21 (18.8) |
Cases with follow-up (N), n (%) |
57 (91.9) |
20 (80.0) |
3 (75.0) |
14 (66.7) |
Sensitivity, % (TP/TP+FN) |
>99.9 (49/ 49) |
>99.9 (18/ 18) |
>99.9 (3/ 3) |
>99.9 (6/ 6) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,594/ 2,602) |
99.0 (203/ 205) |
>99.9 (277/ 377) |
99.8 (3,560/ 3,568) |
PPV, % (TP/N) |
86.0 (49/ 57) |
90.0 (18/ 20) |
>99.9 (3/ 3) |
42.9 (6/ 14) |
Trisomy 13 |
Cases, n (%) |
26 (49.1) |
8 (15.1) |
2 (3.8) |
17 (32.1) |
Cases with follow-up (N), n (%) |
17 (65.4) |
6 (75.0) |
2 (100.0) |
13 (76.5) |
Sensitivity, % (TP/TP+FN) |
>99.9 (10/ 10) |
>99.9 (4/ 4) |
>99.9 (2/ 2) |
>99.9 (7/ 7) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (2,555/ 2,562) |
99.0 (189/ 191) |
>99.9 (276/ 276) |
99.8 (3,561/ 3,567) |
PPV, % (TP/N) |
58.8 (10/ 17) |
66.7 (4/ 6) |
>99.9 (2/ 2) |
53.8 (7/ 13) |
Monosomy X |
Cases, n (%) |
13 (29.5) |
9 (20.5) |
4 (9.1) |
18 (40.9) |
Cases with follow-up (N), n (%) |
9 (69.2) |
7 (77.8) |
3 (75.0) |
13 (72.2) |
Sensitivity, % (TP/TP+FN) |
>99.9 (5/ 5) |
>99.9 (7/ 7) |
>99.9 (3/3) |
>99.9 (6/ 6) |
Specificity, % (TN+TP/TN+TP+FP) |
99.8 (2,550/ 2,554) |
>99.9 (192/ 192) |
>99.9 (277/277) |
99.8 (3,560/ 3,567) |
PPV, % (TP/N) |
55.6 (5/ 9) |
>99.9 (7/ 7) |
>99.9 (3/3) |
46.2 (6/ 13) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 5. Sensitivities, specificities and PPV values for high-risk cases stratified by maternal age
Maternal age, years |
< 25 |
25 – 29 |
30 – 34 |
35 – 39 |
> 40 |
Cases, n (%) |
1,057 (2.5) |
6,889 (16.6) |
16,099 (38.7) |
14,020 (33.7) |
3.541 (8.5) |
Total high-risk cases, n (%) |
8 (0.0) |
41 (0.1) |
125 (0.3) |
220 (0.5) |
151 (0.4) |
Trisomy 21, 18, 13, 13 + 21 |
Cases, n (%) |
6 (1.2) |
36 (7.2) |
103 (20.6) |
207 (41.3) |
149 (29.7) |
Cases with follow-up (N), n (%) |
5 (83.3) |
29 (80.6) |
83 (80.6) |
172 (83.1) |
131 (87.9) |
Sensitivity, % (TP/TP+FN) |
>99.9 (2/ 2) |
>99.9 (22/ 22) |
98.6 (71/ 72) |
>99.9 (160/ 160) |
>99.9 (115/ 115) |
Specificity, % (TN+TP/TN+TP+FP) |
98.4 (180/ 183) |
99.4 (1,190/ 1,197) |
99.5 (2,653/ 2,665) |
99.5 (2,315/ 2,327) |
97.4 (590/ 606) |
PPV, % (TP/N) |
40.0 (2/ 5) |
75.9 (22/ 29) |
85.5 (71/ 83) |
93.0 (160/ 172) |
87.8 (115/ 131) |
Monosomy X |
Cases, n (%) |
2 (0.6) |
5 (1.5) |
22 (6.6) |
13 (3.9) |
2 (0.6) |
Cases with follow-up (N), n (%) |
1 (50.0) |
3 (60.0) |
17 (77.3) |
9 (69.2) |
2 (100.0) |
Sensitivity, % (TP/TP+FN) |
- (1/ 1) |
>99.9 (2/ 2) |
>99.9 (11/ 11) |
>99.9 (5/ 5) |
>99.9 (2/ 2) |
Specificity, % (TN+TP/TN+TP+FP) |
- (179/ 179) |
99.9 (1,170/ 1,171) |
99.8 (2,593/ 2,599) |
99.8 (2,160/ 2,164) |
>99.9 (477/ 477) |
PPV, % (TP/N) |
- (1/ 1) |
66.7 (2/ 3) |
64.7 (11/ 17) |
55.6 (5/ 9) |
>99.9 (2/ 2) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 6. Sensitivities, specificities and PPV values for high-risk cases stratified by gestational age
Gestational age, week+day |
10+0 – 10+6 |
11+0 – 13+6 |
14+0 – 19+6 |
>20+0 |
Cases, n (%) |
7,556 (18.2) |
29,212 (70.2) |
4,392 (10.6) |
447 (1.1) |
Total high-risk cases, n (%) |
104 (1.4) |
371 (4.9) |
48 (0.6) |
22 (0.3) |
Trisomy 21, 18, 13, 13 + 21 |
Cases, n (%) |
93 (18.6) |
345 (68.9) |
44 (8.8) |
19 (3.8) |
Cases with follow-up (N), n (%) |
74 (79.6) |
291 (84.3) |
40 (90.9) |
15 (78.9) |
Sensitivity, % (TP/TP+FN) |
>99.9 (68/ 68) |
99.6 (257/ 258) |
>99.9 (30/ 30) |
>99.9 (15/ 15) |
Specificity, % (TN+TP/TN+TP+FP) |
99.5 (1,248/ 1,254) |
99.3 (4,922/ 4,956) |
98.6 (691/ 701) |
>99.9 (67/ 67) |
PPV, % (TP/N) |
91.9 (68/ 74) |
88.3 (257/ 291) |
75.0 (30/ 40) |
>99.9 (15/ 15) |
Monosomy X |
Cases, n (%) |
11 (25.0) |
26 (59.1) |
4 (9.1) |
3 (6.8) |
Cases with follow-up (N), n (%) |
9 (81.8) |
17 (65.4) |
4 (100.0) |
2 (66.7) |
Sensitivity, % (TP/TP+FN) |
>99.9 (6/ 6) |
>99.9 (12/ 12) |
>99.9 (2/ 2) |
- (1/ 1) |
Specificity, % (TN+TP/TN+TP+FP) |
99.7 (1,186/ 1,189) |
99.9 (4,677/ 4,682) |
99.7 (663/ 665) |
- (53/ 54) |
PPV, % (TP/N) |
66.7 (6/ 9) |
70.6 (12/ 17) |
50.0 (2/ 4) |
- (1/ 2) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 7. Sensitivities, specificities and PPV values for high-risk cases for cases with fetal fractions < 4%, 4%-8%, and > 8%
Fetal fraction, % |
< 4% |
4% – 8% |
> 8% |
Cases, n (%) |
1,015 (2.4) |
17,522 (42.3) |
22,916 (55.3) |
Total high-risk cases, n (%) |
47 (4.6) |
266 (1.5) |
232 (1.0) |
Trisomy 21 |
Cases, n (%) |
19 (5.7) |
148 (44.2) |
168 (50.1) |
Cases with follow-up (N), n (%) |
14 (73.7) |
127 (85.8) |
146 (86.9) |
Sensitivity, % (TP/TP+FN) |
90.9 (10/ 11) |
>99.9 (116/ 116) |
>99.9 (144/ 144) |
Specificity, % (TN+TP/TN+TP+FP) |
97.8 (180/ 184) |
99.6 (3,094/ 3,105) |
99.9 (3,554/ 3,556) |
PPV, % (TP/N) |
71.4 (10/ 14) |
91.3 (116/ 127) |
98.6 (144/ 146) |
Trisomy 18 |
Cases, n (%) |
15 (13.4) |
64 (57.1) |
33 (29.5) |
Cases with follow-up (N), n (%) |
10 (66.7) |
57 (89.1) |
27 (81.8) |
Sensitivity, % (TP/TP+FN) |
>99.9 (7/ 7) |
>99.9 (45/ 45) |
>99.9 (24/ 24) |
Specificity, % (TN+TP/TN+TP+FP) |
98.3 (177/ 180) |
99.6 (3,023/ 3,035) |
99.9 (3,434/ 3,437) |
PPV, % (TP/N) |
70.0 (7/ 10) |
78.9 (45/ 57) |
88.9 (24/ 27) |
Trisomy 13 |
Cases, n (%) |
13 (24.5) |
32 (60.4) |
8 (15.1) |
Cases with follow-up (N), n (%) |
9 (69.2) |
23 (71.9) |
6 (75.0) |
Sensitivity, % (TP/TP+FN) |
>99.9 (6/ 6) |
>99.9 (14/ 14) |
>99.9 (3/ 3) |
Specificity, % (TN+TP/TN+TP+FP) |
98.3 (176/ 179) |
99.7 (2,992/ 3,001) |
99.9 (3,413/ 3,416) |
PPV, % (TP/N) |
66.7 (6/ 9) |
60.9 (14/ 23) |
50.0 (3/ 6) |
Monosomy X |
Cases, n (%) |
0 (0.0) |
22 (50.0) |
22 (50.0) |
Cases with follow-up (N), n (%) |
- |
17 (77.3) |
15 (68.2) |
Sensitivity, % (TP/TP+FN) |
- (0/ 0) |
>99.9 (12/ 12) |
>99.9 (9/ 9) |
Specificity, % (TN+TP/TN+TP+FP) |
- (170/ 170) |
99.8 (2,990/ 2,995) |
99.8 (3,419/ 3,425) |
PPV, % (TP/N) |
- (0/ 0) |
70.6 (12/ 17) |
60.0 (9/ 15) |
n: Number; TP: True positive; FP: False positive; TN: True negative; FN: False negative; N: Cases with obtained fetal outcome/with follow up; unk: Cases without obtained follow up.
Table 8. Relationship between NIPT results and LLR scores/T-Statistics values
|
True positive |
False positive |
|
LLR Score |
T-Statistics value |
LLR Score |
T-Statistics value |
Trisomy 21 |
179.1 ± 9.7 |
16.6 ± 0.5 |
20.9 ± 9.8 |
4.9 ± 0.9 |
Trisomy 18 |
170.5 ± 19.3 |
16.2 ± 1.0 |
44.1 ± 18.0 |
8.0 ± 1.6 |
Trisomy 13 |
126.6 ± 21.7 |
14.6 ± 1.5 |
47.1 ± 17.2 |
8.6 ± 1.6 |
Monosomy X |
50.8 ± 8.5 |
-24.2 ± 2.2 |
48.8 ± 18.9 |
-22.0 ± 4.4 |
Values shown as mean ± standard error of the mean (SEM).

