Introduction
If ontogeny recapitulates phylogeny, than cancerogenesis capitulates ontogeny. Cells in pre-malignant and malignant tumors evolve by natural selection [1-2]. Cancer is a classical example of what evolutionary biologists call multilevel selection: at the level of the organism, cancer is usually fatal so there is selection for genes and the organization of tissues that suppress cancer [3-4]. At the level of the cell, there is selection for increased cell proliferation and survival, such that a future cancer cell will have a competitive advantage over cells that have not acquired hallmarks of cancer [5]. Thus, at the level of the cell there is selection for cancer.
Several types of changes that occur when a cell becomes oncogenic are:
- Cell proliferation, which becomes more similar to that of prokaryotic cells, where cells tend to grow and divide as fast as they can, and the rate of proliferation depends largely on the availability of nutrients in the environment;
- Immortalization, a property of indefinite growth without any other changes in the phenotype, occurs, so as in prokaryotic cells, division is unlimited;
Lloss of contact inhibition occurs, which must be regulated by signals from other cells in the body combined with programs intrinsic to the individual cell. The relation cell-cell are disrupted so that the human cancer cell behaves as an individual prokaryotic cell;
- Metastasis occurs, in which the cancer cell gains the ability to invade normal tissue, move away from the tissue of origin, cross through the walls of the capillary blood vessels, as prokaryotic cells may do, and establish a new colony elsewhere in the human body;
- Huge power consumption is observed, due to the presence of increased ATP consumption in affected cancerogenic cells.
Organisms evolv from ancestral to extant entities have inherited molecular biology pathways for billion of years. In multicellular host, archaeal form of life subvent the host biochemical reactions and induce host cell to provide growth factor for them. The malignant cells emerge as selfish individuals, independent from a cell comunity
All this supports the fact that the biochemical activity of eukaryotic cancer cells is subjected to the influence of prokaryotic gene expression. The cells of a tumor descend from a common ancestral cell that became mutant. There are two classes of genes in which mutations cause transformation: tumor suppressor (antiproliferating) genes and oncogenes. Both of these classes of human genes that cause cancer are of prokaryotic origin. The fact that tumor suppressor genes exist says a lot about evolution and cancerogenesis. This means that the development of modern eukaryotic cells was subjected to the influence and selection of an inherited prokaryotic genomes. The result of this selection is the current human genome that still contains the legacy of the ancestral prokaryotic genome, which is activated in oncogenesis. The oncogenes carried by the DNA viruses specify proteins that inactivate tumor suppressors. The oncogenes carried by retroviruses are derived from cellular genes (proto-oncogenes) and, therefore, may mimic the behavior of gain-of-function mutations and animal proto-oncogenes. In a normal diploid cell, there are two copies of each tumor-suppressor gene, and both copies of the gene must be lost or inactivated to bring about the loss of proliferation control, as a single copy is usually enough for normal regulation of the cell cycle. In contrast, only one copy of a proto-oncogene needs to be mutated into an oncogene to promote cancerogenesis.
On entering the cell, glucose is converted to pyruvate. In normal cell, if oxygene is available, pyruvate undergoes oxidative phosphorylation in mitochondria. If oxygene level are low, pyruvate is converted to lactate in the cytoplasm [6]. Cancer cell, however, drive pyruvate conversion to lactate even in the presence of oxygene. So, that coordination of energy production, both in normal and in cancer cell, between nucleus and mitochondrion can not be separated.
All this suggests that cancer cells behave like prokaryotic cells. Therefore, the metabolism of the tumor cells returns to the evolutionary level overlapping that of the prokaryotic cells. If it is possible to find true nature of cancerogenesis, this would be a prerequisite for finding ways to eradicate the disease of cancer.
Monophyletic origin of the nucleus and mitochondrion
The new theory [7-9] of the origin of eukaryotes is based on the existence and development of a single evolutionary line of cells, the genome of which was the basis for the existence of contemporary cellular compartments (nuclear, mitochondrial, plastids). This gave rise to an endogenous evolution of free-living, independent organisms; i.e., an “archaeal only origin of eukaryotes”.
In prokaryotes, cell division occurs through binary fission, driven by the formation of the septum. Septum formation structurally alters the envelope, and the inner membrane becomes closely connected to the cell wall and outer membrane layer [10]. The network of protein interactions must be carefully tuned, both in time and space, to coordinate the correct, timely generation of two identical daughter cells. Any rearrangement, mutation, or changes in the timing or expression level of genes that participate in septum formation could cause separation of the inner and outer membranes during fission. However, the ftsZ and dnm1-like genes are not necessarily coordinated when they are recruited to the constriction site; this indicates that the inner and outer membrane dividing machineries are not in tight association during the late stage of cell division [11]. It is likely that the addition of new DNA sequences, either in the “mitochondrial” or the “nuclear” replicon, could cause the dramatic fission of the two replicons; this “addition” of new sequences could arise from genome duplication. Dynamin regulates membrane squeezing and peroxisome fission (peroxisomes are organelles surrounded by one membrane [12]. Thus, the inner membrane of the archaeal ancestor of eukaryotes (AAE) might form unique proto-nuclear and proto-mitochondrial outer membranes of the resulting proto-eukaryote (before the evolution of the endomembranes and endoskeleton of the mature modern eukaryote).
The steps proposed during the replication of the duplicated AAE genome, show a mechanism that might have led to the fission of the eukaryotic common ancestral genome into nuclear and mitochondrial compartments (Figure 1). In the final step, after genome fission, invagination of the inner membrane could continue and envelop each of the replicons separately. The nuclear membrane would become an uninterrupted, single, lipid bilayer with an outer face and an inner face, as a vestige of the ancient AAE inner membrane invagination that enclosed the “nuclear" replicon.
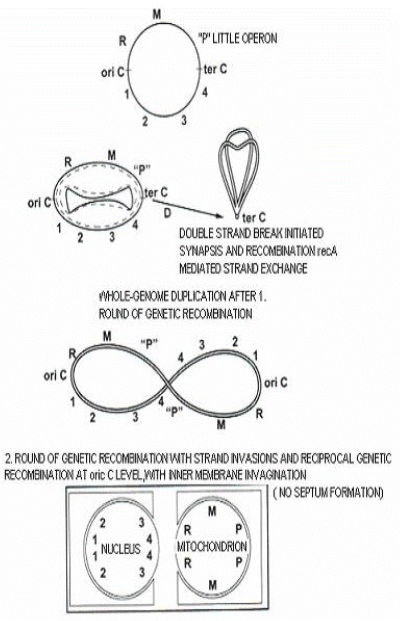
Figure 1. Proposed origin of the eukaryotic nuclear and mitochondrial genomes. The steps proposed show a mechanism that might lead to the fission of the eukaryotic common ancestral genome into nuclear and mitochondrial compartments. 1, 2, 3 and 4 represent different operons in the nucleus; M and R represent different mitochondrial operons, P is the primeval photosynthetic operon.
When direct repeat sequences are near or at the origin of replication, this might lead to functional segregation by genetic recombination, particularly when the maximal capacity of the pragenome has been reached, by whole genome duplication for example. Coupling replication, cdc6 gene activation [13], and pragenome rearrangement could lead to fission between the “nuclear” and “mitochondrial” gene content; this would lead to the formation of the nucleus and mitochondrion.
Figure 1. With genetic recombination, and whole genome duplication of the last eukaryotic common ancestor, till origin of nucleus and mitochondrion.
Archaeal tumor genetics
The best prokariotic candidate for universal ancestor from which eukaryotes emerged, has to be search among archaeal TACK superphylum composed of Thaumarchaeote, Aigarchaeota, Crenarchaeota, and Korarchaeota. Now is possible to account for the presence of Eukaryotic Signature Proteins (ESPs) in TACK lineage. These ESPs-like proteins, including actin, tubulin, dynein, gelsolin, roadblock/lc7-domain, longin, small GTPase, found in Lokiarchaeota [14]. This scenario, esspecially with small GTPase, contrast with previous studies suggesting that small GTPase originated from alpha-proteobacteria progenitor of mitochondria [15]. With this theory there are additional problem, i.e. where are the rest of the alpha-proteobacterial genome goes to; where are the Loki's machinery for energy production, including archaeal ATP synthase? Such advanced machinery should be accompanied bay sofisticated energy production, including oxydative phosphorylation, ATP synthase, NADH dehydrogenese, cytichrome complex, ubiquinone. All this should bi placed in one of the Loki's replicon. Something like in Nitrosomonas faraonicus, for example. Anyway, as it is the case with small GTPase contrasting with previous alfa-protobacterial origin, the same case is with origin of mitochondrion, contrasting with endosymbiotic origin. By the way, in the Loki's article about complex archaea that bridge the gap between prokaryotes and eukaryotes, there is not a words about energy production, the most of the most important biochemical pathway in the cellular life. The fact is that eukaryotic signature biochemistry (ESBs), as spliceosome, the nuclear pore, the endomembrane system, the ubiqutin system (E3 ligase also), the RNAi machinery, the cytosceleton remodeling, the cytosceletal motors, signal transduction, nucleocytoplasmic transport, vesicular trafficing, membrane deformation, cell shape formation, including phagocytosis, are present in the TACK, esspecialy in Lokiarchaeota. Genome of Loki encodes 5381 protein codin genes [predicted by 16], as well as single copies of 16S, and 23S tRNA genes. But, Loki is only one of the new archaeal species that coming up.
All of the eukaryotic genome is transcribed, resulting in numerous non-coding RNA. Micro RNA (miRNA) are small (21-25 nucleotides) non –coding RNA. miRNA processing involved DICER-like protein consisting of RNAse III and helicase domain. Helicase component of DICER protein is specifically related to superfamily II of archaeal helicase, ono of the pivotal events that led to the consolidation of RNAi (RNA interference) in eukaryotic system. Regarding the RNAse III component of the DICER, situatiuon is more complex. Similarity between DROSHA and DICER offer insight of evolution of RNAse III family. Class II RNAse III (DROSHA homolog) may have evolved from Class III RNAse III (DICER homolog) have two tandem RIII Ds in common, suggesting that Class II (DROSHA) might have originated from Class III (DICER) [17]. DICER-like protein is of archaeal origin. In archaea there are miRNA as a powerfull evolutionary machinery, transcription regulation, and defense system against viruses, which is consistent with situation in eukaryotes. Experimental results indicate that miRNA function as a tumor suppressor and oncogenes. Decreased DICER mRNA levels correlate with advanced tumor stage.
A hybrid capable of copying strech of RNA template -100 base long is the first example of a family B polymerase/reverse transcriptase in archaea [18]. Intracellular menadgement of DNA-DNA sequences including reverse transcriptase becomes easy of access. This can eliminated gene complicated gene conversion, and horizontal gene transfer as a evolutionary force in early evolution, and replace with most powerfull mechanism – diversity generating retroelements (DGR). DGR use a process called mutagenic retrohoming for the target replacement of a variable repeat (VR) coding region with a cognate non-coding template repeat RNA. DGR leads to rapid evolution of target protein, and operate in archaeal system. It should not be forgoten that, at the time of monophylet origin of nucleus and mitochondrion, the archaea still retain propriety from RNA world. Together with DGR, WGD, gene duplication, and /or tandemgene duplicationis a major driving evolutionary forces. This create new gene loci with primarly non-existing function by neofunctionalizationa or subfunctionalization. Is it possible that during cancerogenesis, cellular repertoire „remember“ evolutionary pathway leadind back to the origin of the corresponding oncogene or tumor suppressor gene ancestry? [Table 1].
Table 1. List of oncogenes and tumor suppressor genes with archaeal ancestry.
Gene symbol |
Background |
References |
AKT, BCR, BRAF, LCK
PIM, RAF, MAP, RET, ROS,
JUN, ATM, JAK |
ser/thr/tyr kinase |
[19-21] |
ATF, EVI, RUNX1, MAFB,
PLAG, C-JUN, CREB, CEBP,
FOS, TNFA, WT1
|
bZIP |
[22-24]
|
CARD |
caspase |
[25] |
CBL B/C, MDM2, CYLD,
FBXW7, VHL |
Ubiquitin
( with E3) |
[14,26] |
CCND 1,2,3, RAF, CDK |
cyclin dependent
protein kinase |
[14,27,28] |
DDX 5, 6 |
DEAD box RNA helicase |
[29] |
DEK, FUS |
ALBA proteins |
[30] |
FEV, TCF3 |
helix-turn-helix protein |
[31] |
GOLGA, GOPC |
PDZ proteins domain |
[32] |
HMG |
high mobility group prot. |
[33] |
HRAS, KRAS, NRAS, RAS |
small GTPase |
[14] |
MITF, TCF |
helix-loop-helix protein |
[34] |
MLL, KMT2A |
lysine-N-methyltransferase |
[35] |
NCOA4 |
PAS protein motif |
[36] |
NUP, TPR |
nucleoporine |
[37] |
PDGFB, PIK |
phosphatidylinositol synthase |
[38] |
PPARG |
peroxisome |
[39] |
PTPN |
tyrosine phosphatase |
[40] |
SMO |
rhodopsin |
[41] |
MAP |
mitogen-activated protein |
[42] |
USP6 |
cystein protease |
[26] |
BLM, WRN |
Rec Q helicase |
[43] |
CARS |
cysteinil tRNA synthetase |
[44] |
EXT |
exostin |
[45] |
FH |
fumarate hydratase |
[46] |
IDH |
isocitrate dehydrogenase |
[47] |
ARHGEF |
guanine nucleotide
exchange factor |
[48] |
EGFR, ERBBs, FGFR, PDGF
NTRK1, MET, RET, ROS1
|
receptor tyrosine kinase |
++ |
++ The major driving evolutionary force WGD is responsible for origin of receptor tyrosine kinase (RTK) in eukaryotes. RTK possess an extracellular domain composed of EGF and Ig or fibronectin type III domains, a transmembrane domain, and an intracellular tyrosine kinase domain. S-layer proteins of archaea is made up of two domains which are fibronectin III, and Ig-like group 2. Thus, archaea possess all components for RTK assembly. It could be that all RTK family go back to a single common ancestral gene in the archaeal lineage.
Conclusion
The use of cancer cells as a model to study evolution of the eukaryotes (up-down approach) is both interesting and fruitful. Biochemistry and molecular genetics of the cancer cell have sprung to the fore, providing answers as to how the evolution of cancerogenic cells occurred at precisely the time when nuclear and mitochondrial replicons became divided and the eukaryotes appeared.
At this very moment, disordered equilibrium (between membrane basal metabolism, loss of contact inhibition, and aberrant expression of transmembrane genes – future proto-oncogenes) and accelerated cell division (by trigger silenced ancient genes – future proto-oncogenes) had great impact on energy turnover (enormous glycolysis increase). In archaebacterial genome common ancestor sequences for onco-genes and tumor-suppressor genes has already been found. For example, the helicase-associated endonuclease for fork-structured DNA (Hef) is an archaebacterial protein that processes blocked replication forks and participated in tumor-suppressor pathway [49]. Regarding the onco-genes, eukaryotic protein kinase were used to search for eukaryotic-like protein kinase in prokaryotes. This search identified eukaryotic-like protein kinase in archaebacterium Methanococcus vannielii, M. voltae and M. thermolitotrophicus [50]. The archaeabacterial deduced amino acid sequences displayed significant homology with the v-myc gene product and adenovirus E1a oncoprotein [51]. The presence of DNA sequences homologues to the v-myb oncogene was found in both halophilic and methanophilic archaebacteria [52]. RolD protein, plant oncogene product, bears sequence homology with ornithine cyclodeaminase, enzyme of specialized niche archaebacteria [53]. An archaebacterial protein , from Halobacterium halobium, of 84 kD shares common epitopes with the human c-myc protein [54]. Cancer cells of multicellular hosts, operated biochemical pathways that recognizably derived from unicellular ancestor. The descendant heat-shock proteins of thermophilic archaea is now chaperone oncoproteins [55]. All archaea contains chaperons, more similar to the type II chaperons from eukaryotes than to the type I from bacteria, mitochondria and chloroplasts, although some archaea contain type I chaperon [56]. Expanded expression in mammary carcinoma appears to be largely due to the proliferation of overexpressed oncogenes, malfolded muant proteins and that trigger transcriptoin of hsp genes [57]. MCTS1 oncoprotein confers aggressive properties and inhibits apoptosis, and loss of functions in tumor suppressor gene. Using a comparative genomic approach, an ortholog of mcts1 has been identified in archaea [58]. Very interesting study is analysis of domains and domains fusions in human proto-oncogenes [59]. They found that 50% of oncogene domains have their origine in the early stages of evolution, proir to the emergence of metazoans, and no domains are found to arise from mammals.
Genomic, proteomic, and biochemical analysis have reveald the presence of eukaryotic protein kinase (ePK) and phosphatase and an intriguing set of serine-threonine-, and tyrosine-phosphorylated proteins in archaea. A candidate for the direct lineal descendant of the primordial ePK, are found in archaea but absent in bacteria [19]. That means thet first divergence from the LUCA separated the bacterial line of descent from a conjoint eukaryal/archaeal one – primarly via direct inheritance. Cyclin dependent kinase (CDKs) are specific serine/threonine kinase that play an essential role in cell cycle regulation allowing transition between its different phases,and in oncogenesis. The coordinate transition between cell cycle phases depend on family of evolutionarily conserved CDKs present in archaea [60]. From the phylogenetic distribution of protein kinase superfamily, Leonard et al (1998) infer the existence of an ancestral protein kinase prior to the divergence of eukaryotic, bacteria, and archaea [61]. Which is in agreement with hypothesis of endogenous origin of DNA organelles via direct inheritance [7-9], but not as a endosymbiotic event. One of the best example, confirming accuracy of this hypothesis, is a case with Spo11 gene. In all organisms, DNA topoisomerase are essential for untangling chromosomal DNA. The A subunit of archaeal topoisomerase VI is a homologous to the miotic recombination factor Spo11, associated with multiple cancer cell lines [62].
All above indicated that reason for cancerogenesis has to be search among the origin of „archaeal“ tumor suppressor and onco genes whose change cause human disease. In spite of the relation between mutation in the genome of organelles and oncogenesis, there is not a particular organellar gene that trigger malignant transformation. As it is the case with mitochondrial and chloroplast genes, the same is valable for ribosomal genes, havin in mind new upcoming ribosomal DNA organelle [63]. All these facts confirm the accuracy of above hypothesis. Study into gene cluster expression, gene transfer (nucleus ↔ mitochondrion), and genetic recombination study of this transient process, i.e. origin of mitochondrion and nucleus, can provide obvious succession of events in cancerogenesis, which in turn can facilitated diagnostic and treatment of cancer. Looking through the tumor suppressor gene and oncogenes, its archaeal origin profiles through all proteins that are a simplified version of their eukaryotic counterparts. Thus, the basis for oncologicaly provoked suicide of the contemporary archaeal cell, after human death, coming up from expression of the Last Eukaryotic Common Ancestor genes.
References
- Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194: 23-28. [Crossref]
- Merlo LM, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6: 924-935. [Crossref]
- Cairns J (1975) Mutation selection and the natural history of cancer. Nature 255: 197-200. [Crossref]
- Pepper JW, Sprouffske K, Maley CC (2007) Animal cell differentation patterns suppress somatic evolution. PloS Computational Biology 3: e250.
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57-70. [Crossref]
- Kaelin WG Jr, Thompson CB (2010) Q&A: Cancer: clues from cell metabolism. Nature 465: 562-564. [Crossref]
- Stupar M (2008) Archaebacterial whole-genome duplication and origin of Eukaryotes. Nature Preceedings.
- Stupar M (2008b) Nucleogenesis and origin of organelles. Arch Oncol 16: 88-92.
- Stupar M, Vidovic V, Lukac D, Štrbac Lj (2012) Archaebacterial ancestor of eukaryotes and mitochondriogenesis. Biotechnology and Molecular Biology Reviews 7: 84-89.
- de Sousa RF, Gueiros FJ (2001) Maximum likelihood inference of ancestral gene content for the bacterial septosome. Annual International Conference on Intelligent Systems Fortaleza.
- [Crossref] Erickson HP1 (2000) Dynamin and FtsZ. Missing links in mitochondrial and bacterial division. J Cell Biol 148: 1103-1105
- Fujimoto M, Arimura S, Tsutsumi N (2005) Two Arabidopsis dynamin-related proteins 3, DRP3A and DRP3B are involved in mitochondrial and peroxisomal fission ICPMB, Obernai, France.
- Matsunga F, Forterre P, Ishino Y, Myllykallio H (2001) In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc Natl Acad Sci U S A 98: 11152-11157.
- Spang Anja, Saw JH, Jorgensen SI (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173-179.
- Yutin N, Wolf MY, Wolf YI, Koonin EV (2009) The origins of phagocytosis and eukaryogenesis. Biol Direct 4: 9. [Crossref]
- Stupar M, Stafanovic Slavica, Vidovic V (2016) Deciphering principle of life. Schaltungsdienst Langr o.H.G, Berlin, Germany.
- Shabalina SA, Koonin EV (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23: 578-587. [Crossref]
- Jozwiakowski SK, Connolly BA (2011) A modified familyB archaeal DNA Polymerase with reverse transcriptase activity. Chem Biochem 12: 35-37.
- Kennelly PJ (2014) Protein Ser/Thr/Tyr phosphorylation in the Archaea. J Biol Chem 289: 9480-9487. [Crossref]
- Stravopodis DJ, Kyrpides NC (1988) Identification of protein-tyrosine phosphatases in archaea. J Mol Evol 48: 625-627.
- Smith SC, Kennelly PJ, Potts M (1997) Protein-tyrosine phosphorylation in the Archaea. J Bacteriol 179: 2418-2420. [Crossref]
- Ploser Petra and Pfeifer FA (2002) bZIP protein from halophilic archaea: structural features and dimer formation of cGvpE from Halobacterium salinarum. Mol Microbiology 45: 511-520.
- Florence G, Chloe D, Carole J (2013) Solution structure of an archaeal DNA binding protein with an eukaryotic Zink Finger Fold. PLoS ONE 8: e52908.
- Cao J, Shi F, Liu X, Huang G, Zhou M (2010) Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett 584: 4775-4782. [Crossref]
- Bidle KA, Haramat2021 Copyright OAT. All rights reservdence for caspase-like protein expression and activity in the cellular stress response of archaea. Environ Microbiol 12: 1161-1172.
- Julie MFA (2013) Ubiquitin-like proteins and their roles in archaea. Trends Microbiol 21: 31-38.
- Labib K (2010) How do Cdc7 and Cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208-1219.
- Grabowski B, Kelman Z (2001) Autophosphorylation of archaeal Cdc6 homologues is regulated by DNA. J Bacteriol 183: 5459-5464. [Crossref]
- Shimada Y, Fukuda W, Akada Y (2009) Property of cold inducible DEAD-box RNA helicase in hyperthermophilic archeae. Biochem Biophys Res Commun 389: 622-627.
- Richard DJ, Bell SD, White MF (2004) Physical and functional interaction of the archaeal single-stranded DNA-binding protein SSB with RNA polymerase. Nucleic Acids Res 32: 1065-1074.
- Nancy RG, Segovia L, Perez-Rueda E (2011) Diversity and distribution of transcription factors: their partner domains play an important role in regulatory plasticity in bacteria. Microbiology 157: 2308-2318.
- Eichler J, Adams MWW (2005) Posttranslational protein modification in archaea. Microbiol Mol Biol Rev 69: 393-425.
- Murphy FV, Sweet RM, Churchill MEA (1999) The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J 18: 6610-6618.
- Sukront J, Pereira-Leal JB (2016) Are there Rab GTPase in archaea? Mol Biol Evol 33: 1833-1842.
- Niu Y, Xia Y, Wang S (2013) A prototypic lysine methyltransferase 4 from archaea with degenerate sequence specificity methylates chromatin proteins Sul7d and Cren7 in different patterns. J Biol Chem 288: 13728-13740.
- Ponting CP, Aravind L, Schultz J (1999) Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol 289:729-745.
- Neumann Nadja, Lundin D, Poole A (2010) Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PlosOne 5: e13241.
- Jain Samta, Caforio Antonella, Driessen AJM (2014) Biosynthesis of archaeal membrane lipids. Front Microbiol 5: 641-660.
- Gabaldon T, Snel B, Zimmeren F (2006) Origin and evolution of the peroxisomal proteome. Biol Direct 1: 8-21.
- Esser D, Hoffmann Lena, Pham TK (2016) Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiology Reviews 40: 625-647.
- Shabaeva Daria, Galperin MY, Mulkidjanijan AY (2015) Eukaryotic G protein-coupled receptors as descendants of prokaryotic sodium- translocating rhodopsin. Biol Direct 19: 63-80.
- Sedivy JM (2011) Phosphatidylenthanolamine binding protein aka Raf kinase inhibitor protein: A brief history of its discovery and the remarkable diversity of biological functions. For Immunopathol Dis Therap 2: 1-12.
- Fujikane R, Shinagawa H, Ishino Y (2006) The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells 11: 99-110. [Crossref]
- Stathopoulos C, Kim W, Li T, Anderson I, Deutsch B, et al. (2001) Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc Natl Acad Sci U S A 98: 14292-14297. [Crossref]
- Magidovich Hilla, Eichler J (2009) Glycosiltransferases and oligosaccharyltransferases in archaea: putative components of the N-glycosilation pathway in the third domain of life. FEMS Microbiology Letters 300: 122-130.
- Puchegger S, Redl B, Stoffler G (1990) Purification and properties of a thermostable fumarate hydratase from the archaeobacterium Sulfolobus solfataricus. Journal of General Microbiology 136: 1537-1541.
- Spaans SK, Weusthuis RA, van der Oost J, Kengen SW (2015) NADPH-generating systems in bacteria and archaea. Front Microbiol 6: 742. [Crossref]
- Dennis PP (1997) Ancient ciphers: translation in Archaea. Cell 89: 1007-1010. [Crossref]
- Mosedale G, Niedzwiedz W, Alpi A (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nature Structural and Molecular Biology 12: 763-771.
- Smith RF, King KY (1995) Identification of a eukaryotic-like protein kinase gene in Archaebacteria. Protein Sci 4: 126-129. [Crossref]
- Ben-Mahrez K, Sougakoff W, Nakayama M, Kohiyama M (1988) Stimulation of an alpha like DNA polymerase by v-myc related protein of Halobacterium halobium. Arch Microbiol 149: 175-180. [Crossref]
- Perbal B, Kohiyama M (1985) [Existence of sequences homologous to the V-MYB oncogene in the genome of archaebacteria]. C R Acad Sci III 300: 177-180. [Crossref]
- Trovato M, Maras B, Linhares F, Costantino P (2001) The plant oncogene rolD encodes a functional ornithine cyclodeaminase. Proc Natl Acad Sci 90: 13449-13453.
- Ben-Mahrez K, Thierry D, Sorokine I, Danna-Muller A, Kohiyama M et al. (1988) Detection of circulating antibodies against c-myc protein in cancer patients sera. Br J Cancer 57: 529-534.
- Sinkovics JG (2015) The cell survival pathways of the primordial RNA-DNA complex remain conserved in the extant genomes and may function as proto-oncogenes. European J, of Microbiology and Immunology 5: 1.
- Large AT, Goldberg MD, Lundi PA (2009) Chaperons and protein folding in the archaea. Biochem Soc Trans 1: 46-51.
- Calderwood SK, Gong J (2012) Molecular chaperones in mammary cancer growth and breast tumor therapy. J Cell Biochem 113: 1096-1103. [Crossref]
- Hachem A, Nandi S (2007) The oncogene mcts1. Transl Oncogenomics 2: 79-84. [Crossref]
- Qui L (2009) Analysis of domains and domain fusions in human proto-oncogenes. BMC Bioinformatics 10: 88.
- Cavanese Miriam, Santo Loredana, Raje Noopur (2012) Cyclin dependent kinase in cancer: potential for therapeutic intervention. Cancer Biol Ther 13: 451-457.
- Leonard CJ1, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the "eukaryotic" protein kinase superfamily. Genome Res 8: 1038-1047. [Crossref]
- Leonard CJ, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the "eukaryotic" protein kinase superfamily. Genome Res 8:1038-1047.
- Nichols MD, DeAngelis K, Keck JL, Berger JM (1999) Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombinase factor Spo11. EMBO J 18: 6177-6188.
- Stupar M, Vidovic V, Lukac D (2015) Ribosomal DNA intracellular journey and new upcoming ribosomal organelle. Iranian J Science and Technology 39: 267-272.

