A metagenomic approach was used to search for novel nitrile hydrolyzing enzymes from environmental samples. Nitrile compounds are versatile and can be converted into amides, amines, imines, oximes, carboxylic acids, esters and alcohols, encompassing a large group of economically important synthetic intermediates. The pharmaceutical industry particularly requires amides and acids for use as intermediates in the manufacture of many drugs and chemicals. The use of the biodiverse environment in the search for novel catalysts by microbiological selection techniques is the traditional method for the discovery of new enzymes in the development of biocatalysts for different industrial sectors. A fosmid DNA library was prepared from metagenomic DNA extracted from soil samples collected in Ireland. The resulting Escherichia coli clone library was screened using functional selection for nitrile-hydrolyzing enzyme activity using β-hydroxynitriles as substrates; 3-hydroxybutyronitrile, 3-hydroxyglutaronitrile and 3-hydroxy-3- phenylpropionitrile, leading to the identification of 33 clones demonstrating activity. Gene screening of these functionally active isolates for the presence of nitrilase, nitrile hydratase and amidase was performed by conventional PCR and partial gene sequences were identified. Further studies to identify complete gene sequences for cloning and expression are underway towards realizing the commercial potential of the associated enzymes.
Functional metagenomics; Nitrile hydrolysing activity; Nitrilase; Nitrile hydratase; Amidase
The pharmaceutical industry has several motivations to probe the enormous resource that is uncultivated microbial diversity. Presently, there is a global political pressure to encourage industrial/white biotechnology to substantially impact such industrial production. Thus, the development of novel enzymes as the ideal biocatalyst is very desirable, if not required. In fact, metagenomics seems to provide those new molecules with various functions, but eventually, heterologous expression is required for any new enzymes to become an economic success [1].
One interesting example of novel enzymes are the nitrile-degrading enzymes, which by hydrolysis comprises the most common pathway for this microbial metabolism [2]. In this way, we can find two different enzyme systems to work with [3]. The first is by nitrilases, which convert nitriles directly to the corresponding carboxylic acids, via addition of two molecules of water. On the other hand, the second system is a combination of a nitrile hydratase (NHase), which converts the nitrile to amide through a hydrolysis step via the addition of one molecule of water, and an amidase, that subsequently hydrolyzes the amide to the corresponding carboxylic acid via the addition of another molecule of water [4].
The need of amides and acids for use as intermediates in the manufacture of many drugs and chemicals by pharmaceutical industries is great [5]. These may be obtained by traditional chemical methods, but this approach has problems; the conditions required are harsh/extreme and undesirable by-products are produced [3,6,7]. An alternative to the use of traditional chemical methods is the use of nitrile-metabolising enzymes as biocatalysts.
The efficiency of biocatalysts to perform various chemical reactions has been the main attraction for their use in chemical synthesis because of the selectivity presented by natural biocatalysts, such as enantioselectivity (preferential involvement of one enantiomer over the others in a chemical or enzymatic reaction) [8]. In addition, a wide spectrum of chemical compounds that are accepted as xenobiotic substrates for reactions exist, while mild and environmentally sustainable conditions give these biocatalysts some fundamental characteristics for use in different transformations of biotechnological interest [9].
The use of the biodiverse environment in the search for novel catalysts by microbiological selection techniques is the traditional method for discovery of new enzymes in the development of biocatalysts for different industrial sectors. The use of microorganisms has a particular interest due to the short period of cultivation, the wide variety of metabolic processes/enzymes involved and due to an unlimited number of microorganisms in the environment which are very diverse, thus providing the potential discovery of enzymes with many different applications [10].
There are two main different approaches in the isolation of microbial genes/enzymes from the environment; culture-dependent and culture-independent methods. Both allow the characterization of and access to some of the diverse microbiome within a sample and therefore are considered complementary. It has been estimated that 1% of the microorganisms are detected on plates with culture medium due to selective conditions, depending on the composition of the culture media [11,12]. On the other hand, the culture-independent methods indicates a predominance of many uncultivated species [13]. One example of a culture-independent molecular method is metagenomics, which consists of direct extraction of nucleic acids from the environmental sample(s), followed by cloning into a suitable vector, transformation into host cells and screening for the genes and/or functions of interest [14], without going through prior DNA amplification steps.
Currently, uncultured environmental samples can be explored by the metagenomic approach [15]. Most of the well-characterized NHases, amidases and nitrilases, which are largely of bacterial origin, have been obtained by selection methods allowing only the positive strains to grow on a minimum medium with a nitrile as the sole nitrogen source [16-19].
This current study focused on applying functional metagenomics to search for novel nitrile hydrolyzing enzymes using environmental samples. This will allow the cloning, expression and purification of recombinant enzymes, which is the form/stage required by potential industrial partners/customers for screening/scale-up/go-to-market or indeed any industrial exploitation. The nitriles chosen for this study were 3-hydroxybutyronitrile (3HBN), 3-hydroxyglutaronitrile (3HGN) and 3-hydroxy-3-phenylpropionitrile (3HPPN), which are β-hydroxynitriles, which can act as sources of β-hydroxycarboxylic acids by biotransformation - these products could be widely used as chiral precursors for pharmaceutical compounds.
Substrates of interest
Racemic 3HBN and 3HGN were purchased from ENAMINE®, and 3HPPN from Sigma®. All other chemicals were of analytical grade and obtained from Macron Fine Chemicals™ and Acros Organics Chemicals.
Total DNA extraction and metagenomic fosmid library construction
10 soil samples used in this work as sources of metagenomic DNA were obtained from environmental soils collected from terrestrial and aquatic microenvironments in Co. Waterford, Ireland. Total DNA was extracted according to the previously described protocol [20]. The cells were lysed using beads and 20% (x/v) SDS, DNA was then isolated by purification using phenol/chloroform extraction followed by alcohol precipitation. The pellet of DNA was resuspended in TE (10 mM Tris/HCl, 1 mM EDTA, pH 8), treated with RNAse at 37ºC for two hours and stored at -20ºC.
Cloning of metagenomic DNA into the vector pSMART® FOS Vector (Lucigen®) and packaging recombinant lambda phage by Gigapack III XL packaging extract (Stratagene®) were performed as per the manufacturer’s instructions. Briefly, metagenomic DNA samples were subjected to an end-repair reaction to create blunt ends with 5´ phosphate groups for ligation into the blunt, dephosphorylated vector. Fosmids containing the inserts were then packaged with Gigapack III XL and used to infect the Replicator™ FOS strain. Infected cells were spread on YT+CXIS plates (8 g/L bacto-tryptone, 5 g/L yeast extract, 5 g/L NaCl and 15 g/L agar) supplemented with 12.5 µg/mL chloramphenicol, 40 µg/mL X-Gal, 0.4 mM IPTG and 5% (w/v) sucrose, and incubated at 37ºC overnight for selecting transformants. The titer of the packaged fosmid phage particles was first determined to calculate plating requirements. Clones were transferred to 96-well Megablocks® containing LB broth supplemented with the suitable antibiotic and stored after growth at -70ºC in the presence of 20 % (w/v) glycerol.
Fosmid clone selection by functional screening
Individual clones from the metagenomic fosmid library were pre-cultured in 96-well Megablocks® containing 600 µL of TB broth [11.8 g/L bacto-tryptone, 23.6 g/L yeast extract, 9.4 g/L dipotassium hydrogen phosphate (K2HPO4; anhydrous), 2.2 g/L potassium dihydrogen phosphate (KH2PO4; anhydrous), 0.4 % (w/v) glycerol] supplemented with 8 mL filter-sterilized 50 % (w/v) glycerol and 12.5 µg/mL chloramphenicol, and then incubated at 25ºC and 250 rpm for 16 h. After growth, the clones were plated on M9 agar supplemented with 10 mM nitrile substrates and incubated for 6 days at 25ºC for nitrile functional screening. All functional screening in solid medium was carried out in triplicate. For long-term storage, cells were maintained frozen at -70ºC in 20 % (v/v) glycerol.
Gene screening by conventional PCR
NHase, amidase and nitrilase gene screening were performed by direct colony PCR of functional clones. Table 1 shows the combinations of primers used to amplify the NHase and amd genes. The complete α subunit genes were amplified using the forward primer NHA-F and the reverse primer NHA-R. The complete β subunit genes were amplified using the forward primer NHB-F and reverse primer NHB-R. The complete αβ genes were amplified using NHA-F and NHB-R. NHase primers were designed previously [19] in order to amplify Fe-type NHase genes. The complete amidase gene was amplified using the forward primer Amd1-F and the reverse primer Amd1-R [21]. Each 25 µL PCR reaction mixture contained 12.5 µL GoTaq®Green Master Mix (Promega), 10 µM each primer and 15-30 ng DNA or cells adjusted to O.D.@600 = 0.04. The PCR thermal profile consisted of: 1 cycle of 95ºC for 5 min, 30 cycles of 95ºC for 1 min, 56ºC for 1 min, 72ºC for 40 s, followed by 1 cycle of 72ºC for 5 min, with the exception of complete αβ gene amplifications which required an extension time of 1.5 min and of complete amidase gene which required an extension time of 2 min.
Table 1: Primers designed for PCR amplification of the α and β NHase subunit genes, the complete αβ genes for NHase gene, and for the complete amidase gene (amd).
Primers |
Sequences |
Amplicons |
NHA-F |
5’-ATGTCAGTAACGATCGACCAC |
~600 bp |
NHA-R |
5’-AGGCAGTCCTTGGTGA CGAT |
NHB-F |
5’-ATGGATGGAGTACACGATCT |
~600 bp |
NHB-R |
5’-TCAGGCCGCAGG CTCGAGGT |
Amd1-F |
5’-ATACGCGTGAATTCGTGGCGACAATCCGACCTGAC |
~1782 bp |
Amd2-R |
5’-GGTGTTGAGTCGGAGTGGATCTTCGAAACTTCCTAG |
Nitrilases genes were amplified by using degenerate primers and PCR conditions described previously in Coffey et al. [19].
Metagenomic library screening for clones expressing nitrile metabolizing activity
To identify novel genes coding for nitrile hydrolyzing enzymes from new sources, a fosmid metagenomic library was constructed with total DNA isolated from environmental samples collected in Ireland from terrestrial and aquatic microenvironments. A total of 1,2 x 104 clones was obtained from the metagenomic library. These clones were individually screened for nitrile activity on solid medium by functional screening, indicated by growth using the selected nitrile as the sole source of nitrogen. Of the 1.2 x 104 clones screened, 16, 12 and 5 clones showed activity towards 3-hydroxyglutaronitrile, 3-hydroxybutyronitrile, and 3-hydroxy-3-phenylpropionitrile respectively.
PCR screening
A total of 33 clones were confirmed for the ability to use nitriles as the sole nitrogen source and were submitted to gene screening using conventional PCR. PCR amplification with the amidase primers is shown in Figure 1. All the results of the DNA fragments amplifications are shown in Table 2.
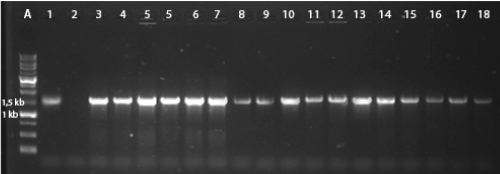
Figure 1. A total of 33 clones were confirmed for the ability to use nitriles as the sole nitrogen source and were submitted to gene screening using conventional PCR. PCR amplification with the amidase primers.
Table 2. Numbers of clones yielding PCR products of the expected size of nitrile hydrolyzing genes.
Functional screening |
Amidase |
Nitrilase |
NHase |
Clones from 3HPPN |
6 |
19 |
- |
Clones from 3HBN |
10 |
39 |
- |
Clones from 3HGN |
_ |
25 |
- |
To construct a metagenomic library to search for new nitrile-metabolizing enzymes, we used DNA fragments representing a mixed microbial soil population obtained in Ireland. A total of 10 environmental samples collected in Ireland from terrestrial and aquatic microenvironments were processed into genomic DNA libraries containing fragments approximately 38-40 kb in size.
We chose a function-based metagenomic approach, which allows the identification of novel nitrile metabolizing enzymes, making it possible to focus gene screening and therefore detect only enzymes that are functional [22].
It is a common practice for enhancing the desired functions in a microbial community to induce the growth of specific microorganisms by applying selective enrichment methods to the sample to produce an increased screening hit rate [23-28]. For instance, Grant et al. [29] used metagenomic DNA of cultures grown in medium containing carboxymethylcellulose as the only carbon source, and they observed that the number of glycosyl hydrolases detected was about four times greater than the number identified in metagenomic libraries obtained using DNA taken directly from environmental samples.
A total of 1.2 x 104 clones was obtained from the metagenomic library. The 33 positive clones demonstrating growth on the chosen nitriles were subjected to conventional PCR screening. Our results demonstrated that 48 % of the positive clones contain the complete amidase gene. In contrast, no clones have presented NHase genes of the sequence type targeted by the chosen primers (Fe-type NHase genes with similar homology to those often seen in Rhodococci, such as Rhodococcus erythropolis strain AJ270 (accession number AJ490527) or strain N771 (accession number AJ716152). It is important to point out that these clones may have a different NHase that the primers do not amplify due to sequence divergence at the primer binding sites. On the other hand, most of them yielded potential PCR products for a nitrilase gene.
In summary, this study consisted of applying functional metagenomics to search for novel nitrile hydrolyzing enzymes using environmental samples collected in Ireland. Gene screening of the positive clones demonstrating the potential presence of nitrilase and amidase genes. Further work to identify complete gene sequences for cloning and expression is underway towards realizing commercial potential. We believe that by using this molecular approach, we may build an extraordinary bank of clones producing novel nitrile-metabolizing enzymes.
We thank CAPES (Coordination for the Improvement of Higher Education Personnel) for the scholarship and financial support.
Compliance with ethical standards
Not applicable.
Conflict of interest
The authors declare that there is no conflict of interest of a scientific or commercial nature. The authors have no relevant affiliations to, or financial support from any organization that may have a financial interest in the subject matter.
- Prophages S (2005) Metagenomics and industrial applications. 3: 510-516. [Crossref]
- Banerjee A, Sharma R, Banerjee UC (2002) The nitrile degrading enzymes: current status and future prospects. APPl. Microbiol. Biorechnol 60:33-44.
- Crabtree RH (2009) Handbook of Green Catalysis: Biocatalysis.
2021 Copyright OAT. All rights reserv
- Kiełbasiński P, Rachwalski M, Mikołajczyk M, Rutjes FPJT (2008) Nitrilase-catalysed hydrolysis of cyanomethyl p-tolyl sulfoxide: stereochemistry and mechanism. Tetrahedron: Asymmetry 19: 562–567.
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M et al. (2001) Industrial biocatalysis today and tomorrow. Nature 409:258–268. [Crossref]
- Johannes TW, Simurdiak MR, Zhao H (2006) Biocatalysis. Encycl. Chem. Process. doi:10.1081/E-ECHP-120017565.
- Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM et al. (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb. Cell Fact. 11:142. [Crossref]
- Silverman RB (2002) The organic chemistry of enzyme-catalyzed reactions.
- Faber K (2011) Biotransformations in Organic Chemistry. Springer-Verlag.
- Adrio JL, Demain AL (2014) Microbial enzymes: tools for biotechnological processes. Biomolecules 4: 117–39.
- Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95: 6578–6583.
- Torsvik V, Goksøyr J, Daae FL, Torsvik V, Goksyr J et al. (1990) High diversity in DNA of soil bacteria. Appl. Environ. Microbiol., 56: 782–787. [Crossref]
- Schloss PD, Larget BR, Handelsman J (2004) Integration of Microbial Ecology and Statistics: a Test To Compare Gene Libraries. Appl. Environ. Microbiol. 70: 5485–5492. [Crossref]
- Handelsman J (2004) Metagenomics : Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev. 68: 669–685.
- Daniel R (2005) The metagenomics of soil. Nat. Rev. Microbiol. 3: 470-8. [Crossref]
- Martínková L, Vejvoda V, Kaplan O, Křen V, Bezouška K et al. (2008) Nitrilases from Filamentous Fungi. In Modern Biocatalysis. . Wiley-VCH Verlag GmbH & Co. KGaA 229–245.
- Coady TM, Coffey LV, O‘Reilly C, Owens EB, Lennon CM (2013) A high throughput screening strategy for the assessment of nitrile-hydrolyzing activity towards the production of enantiopure β-hydroxy acids. J. Mol. Catal. B Enzym. 97: 150–155.
- Coffey L, Clarke A, Duggan P, Tambling K, Horgan S et al. (2009) Isolation of identical nitrilase genes from multiple bacterial strains and real-time PCR detection of the genes from soils provide evidence of horizontal gene transfer. Arch. Microbiol. 191: 761–71.
- Coffey L, Owens E, Tambling K, O’Neill D, O’Connor L et al. (2010) Real-time PCR detection of Fe-type nitrile hydratase genes from environmental isolates suggests horizontal gene transfer between multiple genera. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 98: 455–463.
- Stevenson DM, Weimer PJ (2007) Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. [Internet]. Appl. Microbiol. Biotechnol. 75: 165–74.
- Brandão PFB, Clapp JP, Bull AT (2003) Diversity of Nitrile Hydratase and Amidase Enzyme Genes in Rhodococcus erythropolis Recovered from Geographically Distinct Habitats Diversity of Nitrile Hydratase and Amidase Enzyme Genes in Rhodococcus erythropolis Recovered from Geographically Distinct. Appl. Environ. Microbiol.
- Sleator RD, Shortall C, Hill C (2008) Metagenomics. [Internet]. Lett. Appl. Microbiol 47: 361–6. [Crossref]
- Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 77: 1153–61. [Crossref]
- Uchiyama T, Miyazaki K (2009) Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol 20: 616–22.
- Rabausch U, Juergensen J, Ilmberger N, Böhnke S, Fischer S et al. (2013) Functional screening of metagenome and genome libraries for detection of novel flavonoid-modifying enzymes. Appl. Environ. Microbiol. 79: 4551–4563.
- Michael GB, Simoneti R, Da Costa M, Cardoso M (2003) Comparison of different selective enrichment steps to isolate Salmonella sp. from feces of finishing swine. Brazilian J. Microbiol. 34: 138–142.
- Lam KN, Cheng J, Engel K, Neufeld JD, Charles TC (2015) Current and future resources for functional metagenomics. Front. Microbiol. 2015, 6:1–8.
- Wang K, Wang S, Huang R, Liu Y (2012) Advances of metagenomics in discovering novel biocatalysts [Internet]. Chin. J. Biotechnol. 28: 420–31. [Crossref]
- Grant S, Sorokin DY, Grant WD, Jones BE, Heaphy S (2004) A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures. Extremophiles 8: 421–429.

