Purpose: Anorexia nervosa (AN) is a serious psychiatric disorder characterized by abnormal eating behaviors, resulting in weight loss and increased mortality. Although more common in females, an estimated 5 to 10% of affected patients are males. Up to now, the exact cause of male AN is unknown. As with many psychiatric diseases, it's probably a combination of genetic, biological, psychological and environmental factors. Here, we used whole-genome bisulfite sequencing to determine the methylome of male individuals with AN.
Methods: We analyzed by bisulfite sequencing 3,340,894 biologically relevant CpG sites (Illumina TruSeqMethyl Capture EPIC kit) of 6 male patients affected with AN restrictive type. To reduce the environment effect, 4 related unaffected individuals were selected as controls.
Results: Comparisons between male patients affected with AN restrictive type and unaffected controls showed 153 differentially methylated regions and 1812 differentially methylated CpGs that corresponded to genes relevant to metabolic and nutritional status, psychiatric status and immune function. Moreover, the String network analysis software identified a subnetwork, related to MAPK signaling pathway, PI3K-Akt signaling pathway and neurotrophin signaling pathway.
Conclusions: Our findings replicate several results concerning several target genes such as PRKAG2, RPTOR, and ICAM5 previously identified in female AN, and identified novel signaling pathways involving PI3K-Akt and neurotrophin signaling pathway disturbed in AN.
anorexia nervosa, restrictive type, anorexia, whole-genome bisulfite sequencing
Anorexia nervosa (AN) is a complex neuropsychiatric disorder characterized by weight loss, and an intense fear of gaining weight. Family and twin studies of AN have shown that genetic and environmental factors play important roles in the pathogenesis of AN. Twin studies have estimated the heritability to be~56% [1]. However, while several genes were identified by candidate gene studies and genome-wide association studies (GWAS), they often failed to replicate in other studies [2-5]. The same observation was found for major depression sharing common genetic and environmental risk factors with AN [6]. Indeed, young people who suffered from AN are at high risk of depression [7]. Epigenetics and environmental factors might play a crucial role in the development of AN as well as in major depressive disorder [8]. Recently, a genome-wide DNA methylation study was conducted in women with active AN, AN in remission and in non-AN controls. Global analysis of the 3 groups revealed 295 differentially methylated sites (DMS) representing 277 genes. Some of the identified genes were related to nutrition, to bone and tissue heath, immune function/inflammation, as well as general or transcriptional processes, and glia-neuron interaction [9].
As AN is nine times more often in females than in males, focusing on gender specificities might help to shed light on its overall nature. We therefore propose an analysis of several male AN patients, supporting in part the recent findings and identifying a novel disease-relevant pathway.
We analyzed more than 3.3 M biologically relevant CpGs (Illumina TruSeqMethyl Capture EPIC kit) of 6 male patients affected with AN restrictive type. This approach captures 3,340,894 CpG sites including 26,981 CpG islands (107 Mb), including American College of Medical Genetics (ACMG) genes, coding genes known to be involved in cancer, coding exons from Ensemble 70, and 100 promoters defined as being of high interest and difficult to sequence. To reduce the environment effect, 4 related unaffected individuals- parents of two AN individuals - were selected as controls. Briefly, libraries were prepared according to manufacturer protocol (using the TrueSeq DNA Methylation Kit (cat. EGMK81312, Illumina Inc., San Diego, CA), the NEXTflex Bisulfite Sequencing Kit (cat. 5119–02) and the 24 NEXTflex Bisulfite Sequencing Barcodes (cat. 511913)). After PCR-amplification, the bisulfite-treated Libraries were clustered on a V3 paired-end read flow cell and sequenced for 100 cycles on an Illumina NextSeq500 System (Illumina). Fastq files were generated using Illumina’s software CASAVA v1.8.2. Raw fastq reads were processed by a custom pipeline that consists of: (i) filtering raw fastq reads for pass filter reads, (ii) trimming adapter sequence by Trim Galore, (iii) genomic alignments performed using Bismark to reference human genome hg19, and (iv) methylation calling by a Bismark script (https://github.com/nf-core/methylseq). The R package methyKit was used for analysis and annotation of DNA [10]. After adjustment for potential confounders and exclusion of age-related CpG, differences were shown between AN individuals and controls. We initially investigated the differentially methylated regions (DMRs) in AN using a computational algorithm MethylKit with a stringent statistical cutoff of q-value (FDR adjusted p-value<0.01) and a minimum 25% change in methylation between AN patients and controls.
This approach allowed us to identify a total of 153 DMRs in AN (Table 1). Of these, 53 DMRs were hypomethylated and 100 were hypermethylated, indicating that more genic regions tended to be methylated in AN individuals, similar to previous observations [9]. We further examined the genomic distribution of differentially methylated CpGs (DMCs). A total of 1812 DMC sites were identified in patients with AN compared to controls (Supplementary Table 1). Among the 1812 DMCs, 748 were hypomethylated, while 1065 were hypermethylated in the AN samples. Among these 1812 DMCs, 216functional genic regions included at least 2 DMCs. Interestingly, when we compared our DMCs list with the short list of Steiger and colleagues, 32 genes were found in both lists (Supplementary Table 2) [9]. Moreover, among the 100 top-ranked differentially methylated positions identified in patients with major depressive disorder, six were also found in our study (GNG4, SLC39A12, CRTAC1, MYO7A, NTM and RMST), four of them playing a role in neurite extension and neurogenesis (GNG4, SLC39A12, NTM and RMST) [8].
Table 1. Identification of differentially methylated regions (DMR) was performed using the predefined regions of Illumina TruSeq Methylcapture EPIC kit presenting at least 25 % methylation difference between ANcases and controls (column Change) with a q-value (FDR corrected p-value) threshold of 0.01
DMR |
Chr |
Nb Genes |
Genes |
p value |
q value |
Change |
chrX_114502896_114503051 |
chrX |
0 |
- |
2.22451E-15 |
3.71584E-14 |
59.78484933 |
chr14_54375093_54375328 |
chr14 |
1 |
AL138479. |
2.65207E-24 |
1.18277E-22 |
48.00682879 |
chr8_80036078_80036321 |
chr8 |
0 |
- |
2.44128E-36 |
2.55548E-34 |
46.08190008 |
chr11_79289117_79289197 |
chr11 |
0 |
- |
2.09678E-48 |
3.89352E-46 |
44.01889299 |
chr2_165342265_165342368 |
chr2 |
1 |
GRB14 |
5.17941E-11 |
4.44995E-10 |
43.62139918 |
chr5_118811413_118811668 |
chr5 |
2 |
HSD17B4|snoU1 |
4.63731E-11 |
4.01928E-10 |
43.59922516 |
chr19_45113900_45114110 |
chr19 |
2 |
CEACAM22P|IGSF23 |
9.32156E-32 |
7.39337E-30 |
40.63822929 |
chr12_39885082_39885307 |
chr12 |
0 |
- |
1.48466E-54 |
3.57503E-52 |
40.49521423 |
chr11_99564881_99564954 |
chr11 |
1 |
CNTN5 |
9.37129E-15 |
1.42916E-13 |
39.62334329 |
chr20_46228572_46228783 |
chr20 |
1 |
NCOA3 |
1.11712E-13 |
1.47266E-12 |
39.56217898 |
chr12_9507242_9507307 |
chr12 |
0 |
- |
1.05726E-24 |
4.90641E-23 |
38.46786372 |
chr2_238213149_238213414 |
chr2 |
0 |
- |
1.58339E-06 |
5.42033E-06 |
37.58090232 |
chr9_88891303_88891317 |
chr9 |
1 |
ISCA1 |
1.08415E-06 |
3.8617E-06 |
37.31409719 |
chr7_134279040_134279370 |
chr7 |
0 |
- |
7.42127E-19 |
1.90865E-17 |
36.8404498 |
chr11_125477842_125477944 |
chr11 |
1 |
STT3A |
1.37342E-52 |
3.02928E-50 |
36.14363165 |
chr4_85873367_85873551 |
chr4 |
1 |
WDFY3 |
8.771E-29 |
5.54172E-27 |
35.86458464 |
chr1_228243499_228243750 |
chr1 |
1 |
WNT3A |
7.15126E-21 |
2.28614E-19 |
35.584194 |
chr2_131046231_131046636 |
chr2 |
5 |
MTND4P2|MTND5P2|MTND6P|AC068137.1|AC068137.1 |
1.32673E-40 |
1.75348E-38 |
35.46103678 |
chr7_39723763_39723940 |
chr7 |
1 |
RALA |
2.01448E-11 |
1.85339E-10 |
34.0085943 |
chr6_132297421_132297621 |
chr6 |
1 |
RP11-69I8. |
0.001384883 |
0.002169711 |
33.37310303 |
chr3_108880309_108880502 |
chr3 |
0 |
- |
7.64309E-11 |
6.36725E-10 |
33.06601128 |
chr1_66753890_66754101 |
chr1 |
1 |
PDE4B |
0.00182074 |
0.00275054 |
32.96104032 |
chr2_66654487_66654810 |
chr2 |
2 |
MEIS1-AS3|MEIS1 |
2.10022E-69 |
7.72486E-67 |
32.72071839 |
chr5_97764439_97764614 |
chr5 |
0 |
- |
0.000291866 |
0.000557559 |
32.43193392 |
chr9_115957266_115957508 |
chr9 |
1 |
FKBP15 |
1.69011E-15 |
2.87555E-14 |
32.29050602 |
chr15_50209373_50209588 |
chr15 |
1 |
ATP8B4 |
3.01371E-25 |
1.45926E-23 |
31.98261372 |
chr17_11608628_11608839 |
chr17 |
1 |
DNAH9 |
6.97091E-30 |
4.8207E-28 |
31.79815222 |
chr4_3040168_3040507 |
chr4 |
2 |
GRK4|RNU6-204 |
7.3598E-14 |
9.96918E-13 |
31.65690685 |
chr6_88956453_88956584 |
chr6 |
0 |
- |
9.99041E-12 |
9.68676E-11 |
31.54201058 |
chr21_15436054_15437354 |
chr21 |
3 |
AP001347.|ANKRD20A18|RNA5SP48 |
0 |
0 |
31.16688815 |
chr3_49712243_49712522 |
chr3 |
4 |
BSN|APEH|MST1|AC099668. |
8.07314E-25 |
3.79115E-23 |
31.03180163 |
chr12_25264152_25264371 |
chr12 |
2 |
LRMP|CASC1 |
8.78331E-27 |
4.8313E-25 |
30.29058703 |
chrX_9121569_9121778 |
chrX |
1 |
FAM9B |
0.000426217 |
0.000777831 |
30.08088109 |
chr3_84932517_84932772 |
chr3 |
2 |
LINC00971|LINC02025 |
2.07283E-18 |
5.07715E-17 |
29.94169954 |
chr7_123928236_123928461 |
chr7 |
2 |
RP5-921G16.|RP11-264K23. |
3.518E-31 |
2.67311E-29 |
29.91216787 |
chr9_106087270_106087465 |
chr9 |
1 |
LINC01492 |
8.36825E-19 |
2.13729E-17 |
29.78985686 |
chr5_120388958_120389191 |
chr5 |
2 |
AC008565.|CTD-2613O8. |
7.61105E-44 |
1.17667E-41 |
29.58350371 |
chr11_8927411_8927650 |
chr11 |
2 |
ST5|AKIP1 |
4.49366E-10 |
3.25802E-09 |
29.52623011 |
chr6_159572123_159572413 |
chr6 |
0 |
- |
7.42108E-39 |
8.88692E-37 |
29.31934038 |
chr22_49625975_49626187 |
chr22 |
0 |
- |
2.92003E-11 |
2.61571E-10 |
29.27879223 |
chr12_9776968_9777167 |
chr12 |
2 |
LOC374443|RNU6-700 |
1.12651E-09 |
7.58502E-09 |
29.08923252 |
chr3_170332310_170332590 |
chr3 |
1 |
SLC7A14-AS1 |
1.8717E-08 |
9.81278E-08 |
29.01098901 |
chr4_77986307_77986536 |
chr4 |
1 |
CCNI |
3.91084E-14 |
5.48555E-13 |
28.97085991 |
chr22_42176013_42176129 |
chr22 |
1 |
MEI1 |
0.000206038 |
0.000410706 |
28.81962113 |
chr10_25240859_25240994 |
chr10 |
2 |
PRTFDC1|RP11-165A20. |
6.26124E-28 |
3.71906E-26 |
28.60587752 |
chr3_79966821_79966915 |
chr3 |
0 |
- |
1.15638E-10 |
9.34284E-10 |
28.60217066 |
chr3_45649141_45649463 |
chr3 |
1 |
LIMD1 |
6.28165E-62 |
1.8899E-59 |
28.50314686 |
chr6_25166613_25166879 |
chr6 |
1 |
CMAHP |
2.17351E-96 |
1.46968E-93 |
28.37986243 |
chr3_152213629_152213846 |
chr3 |
1 |
RP11-362A9. |
6.8342E-05 |
0.000155335 |
28.26746276 |
chr11_59836664_59836958 |
chr11 |
2 |
MS4A3|RP11-736I10. |
3.63343E-09 |
2.21257E-08 |
28.0516934 |
chr20_14124320_14124383 |
chr20 |
1 |
MACROD2 |
1.40341E-20 |
4.35847E-19 |
28.02413196 |
chr22_32728242_32728474 |
chr22 |
1 |
RP1-149A16.1 |
8.85421E-11 |
7.29047E-10 |
27.91646442 |
chr2_183106034_183106226 |
chr2 |
1 |
PDE1A |
9.65506E-09 |
5.3791E-08 |
27.85254866 |
chr6_118401762_118401938 |
chr6 |
2 |
SLC35F1|LOC105377967 |
4.64459E-19 |
1.22408E-17 |
27.79487179 |
chr10_91369814_91369992 |
chr10 |
1 |
PANK1 |
1.57036E-21 |
5.3785E-20 |
27.77056277 |
chr12_49121182_49121257 |
chr12 |
1 |
TEX49 |
8.24722E-13 |
9.54004E-12 |
27.60425962 |
chr5_95362159_95362388 |
chr5 |
1 |
LOC101929710 |
4.61645E-19 |
1.21738E-17 |
27.51574796 |
chr17_16935456_16935522 |
chr17 |
0 |
- |
4.58885E-09 |
2.74027E-08 |
27.46293683 |
chr3_42093299_42093629 |
chr3 |
2 |
TRAK1|RP11-193I22. |
6.20321E-37 |
6.69223E-35 |
27.41943731 |
chr3_149867507_149867632 |
chr3 |
1 |
LOC105374313 |
3.07268E-11 |
2.74327E-10 |
27.29693742 |
chrX_43503885_43504133 |
chrX |
0 |
- |
4.40393E-09 |
2.64009E-08 |
27.18926273 |
chr14_77035752_77035960 |
chr14 |
1 |
RP11-187O7. |
6.2288E-12 |
6.23127E-11 |
27.15821169 |
chr10_52771069_52771259 |
chr10 |
1 |
PRKG1 |
3.87096E-16 |
7.14936E-15 |
27.04932754 |
chr2_38055293_38055466 |
chr2 |
1 |
LINC00211 |
7.48568E-28 |
4.4228E-26 |
27.03032591 |
chr1_108245511_108245769 |
chr1 |
1 |
VAV3 |
3.17027E-12 |
3.3165E-11 |
26.94594044 |
chr11_132891631_132891819 |
chr11 |
1 |
OPCML |
4.6077E-05 |
0.000109674 |
26.80024478 |
chr8_8538073_8538307 |
chr8 |
0 |
- |
6.60262E-14 |
8.99085E-13 |
26.64249158 |
chr8_107757803_107757980 |
chr8 |
1 |
OXR1 |
0.000716855 |
0.001224528 |
26.62186488 |
chr21_36250641_36250940 |
chr21 |
2 |
LOC100506403|RUNX1 |
3.11176E-25 |
1.50565E-23 |
26.55673748 |
chr9_14910433_14910528 |
chr9 |
1 |
FREM1 |
8.33159E-21 |
2.64892E-19 |
26.53151371 |
chr3_10851357_10851586 |
chr3 |
1 |
SLC6A11 |
0.004410223 |
0.005890646 |
26.40737551 |
chr9_73216788_73217020 |
chr9 |
1 |
TRPM3 |
3.0921E-15 |
5.06212E-14 |
26.40133464 |
chr17_77906666_77906803 |
chr17 |
3 |
LINC01979|LINC01978|TBC1D16 |
3.90756E-08 |
1.9197E-07 |
26.38634157 |
chr15_101507155_101507328 |
chr15 |
1 |
LRRK1 |
5.16927E-09 |
3.05155E-08 |
26.37814064 |
chr10_87988149_87988359 |
chr10 |
1 |
GRID1 |
0.003497861 |
0.004828383 |
26.35618749 |
chr13_46978725_46978934 |
chr13 |
2 |
RUBCNL|RNU6-68 |
2.29746E-12 |
2.46329E-11 |
26.04182226 |
chrX_149197068_149197285 |
chrX |
1 |
LINC00894 |
2.44211E-07 |
1.01031E-06 |
26.00779648 |
chr16_66453623_66453908 |
chr16 |
2 |
LINC00920|BEAN1 |
3.68957E-06 |
1.15579E-05 |
25.85630262 |
chr1_165186330_165186523 |
chr1 |
3 |
LMX1A|RP11-38C18.|RP11-38C18. |
0.000275984 |
0.000530947 |
25.72822521 |
chr12_6662473_6662654 |
chr12 |
3 |
IFFO1|RP5-940J5.|NOP2 |
9.5204E-29 |
6.00387E-27 |
25.71991419 |
chr4_129308317_129308548 |
chr4 |
1 |
LINC02615 |
2.02201E-09 |
1.29361E-08 |
25.68972744 |
chr4_102756899_102757268 |
chr4 |
1 |
BANK1 |
8.27941E-46 |
1.39081E-43 |
25.66642434 |
chr6_101852800_101852969 |
chr6 |
1 |
GRIK2 |
1.14283E-19 |
3.23959E-18 |
25.50742035 |
chr7_147500652_147500831 |
chr7 |
1 |
CNTNAP2 |
4.69478E-06 |
1.43535E-05 |
25.47112462 |
chr2_130683180_130683508 |
chr2 |
2 |
PLAC9P1|LINC01856 |
2.6716E-16 |
5.03565E-15 |
25.46823547 |
chr3_69157750_69157970 |
chr3 |
2 |
ARL6IP5|LMOD3 |
6.39788E-06 |
1.89017E-05 |
25.46542143 |
chr12_104676677_104676883 |
chr12 |
2 |
TXNRD1|RP11-818F20. |
4.32249E-15 |
6.92567E-14 |
25.4273867 |
chr9_4375797_4376098 |
chr9 |
1 |
AL162419. |
3.07166E-06 |
9.81114E-06 |
25.41113806 |
chr6_55377549_55377764 |
chr6 |
1 |
HMGCLL1 |
4.38355E-45 |
7.21874E-43 |
25.36445879 |
chr8_80816076_80816210 |
chr8 |
0 |
- |
2.50289E-06 |
8.16528E-06 |
25.34385329 |
chr5_171810643_171810847 |
chr5 |
1 |
SH3PXD2B |
2.88223E-10 |
2.16508E-09 |
25.2488391 |
chr2_66648614_66648933 |
chr2 |
1 |
MEIS1-AS3 |
1.28929E-22 |
4.90242E-21 |
25.21383356 |
chr14_96583490_96583604 |
chr14 |
0 |
- |
0.003078364 |
0.004325989 |
25.19935377 |
chr22_16864741_16864893 |
chr22 |
1 |
ABCD1P |
0.002259786 |
0.003313591 |
25.19298246 |
chr10_90483503_90483709 |
chr10 |
2 |
LIPK|KRT8P3 |
3.47526E-44 |
5.45461E-42 |
25.15294597 |
chr6_7699982_7700169 |
chr6 |
0 |
- |
1.47903E-12 |
1.64125E-11 |
25.12462285 |
chr3_195501977_195502276 |
chr3 |
1 |
MUC4 |
1.31243E-47 |
2.3649E-45 |
25.08813062 |
chr18_29303346_29303538 |
chr18 |
3 |
RN7SKP4|LRRC37A7|RP11-549B18. |
1.62011E-27 |
9.3655E-26 |
25.03570419 |
chr3_95424692_95425326 |
chr3 |
0 |
- |
3.3968E-14 |
4.80683E-13 |
25.03516155 |
chr3_180102299_180102384 |
chr3 |
0 |
- |
9.13491E-07 |
3.3066E-06 |
25.01885857 |
chr22_35587422_35587713 |
chr22 |
2 |
LINC01399|COX7BP |
1.08406E-10 |
8.79565E-10 |
-25.01400968 |
chr20_36132841_36133069 |
chr20 |
1 |
BLCAP |
1.55091E-18 |
3.84306E-17 |
-25.14666861 |
chr19_47082930_47083060 |
chr19 |
2 |
PPP5D1|AC011551. |
6.29188E-21 |
2.02349E-19 |
-25.24712874 |
chr19_49572906_49573157 |
chr19 |
4 |
NTF4|CTB-60B18.1|CTB-60B18.1|KCNA7 |
5.89203E-07 |
2.23171E-06 |
-25.34019384 |
chr3_11489044_11489192 |
chr3 |
1 |
ATG7 |
7.90562E-10 |
5.47842E-09 |
-25.40198715 |
chr17_75631717_75631979 |
chr17 |
0 |
- |
1.84137E-05 |
4.85349E-05 |
-25.53803605 |
chr1_19176711_19177105 |
chr1 |
2 |
TAS1R2|RP13-279N23. |
0.000375134 |
0.000695164 |
-25.58525242 |
chr11_5840963_5841191 |
chr11 |
2 |
TRIM5|OR52N2 |
2.88714E-66 |
9.68772E-64 |
-25.69185678 |
chr2_55378165_55378300 |
chr2 |
0 |
- |
9.89458E-19 |
2.51065E-17 |
-25.78431373 |
chr13_19315890_19316198 |
chr13 |
3 |
ZNF965|LINC00417|CYP4F34 |
8.90044E-14 |
1.19079E-12 |
-25.87364383 |
chr18_61669993_61670289 |
chr18 |
1 |
SERPINB8 |
8.41432E-63 |
2.5838E-60 |
-26.04262437 |
chr20_32885922_32886673 |
chr20 |
1 |
AHCY |
2.8332E-44 |
4.47838E-42 |
-26.45197851 |
chr6_106252261_106252558 |
chr6 |
0 |
- |
6.2211E-107 |
4.9875E-104 |
-26.54101932 |
chr2_25383849_25384399 |
chr2 |
3 |
EFR3B|RP11-509E16|POMC |
4.25503E-85 |
2.26062E-82 |
-26.70759651 |
chr4_131633369_131633569 |
chr4 |
0 |
- |
1.21778E-09 |
8.14225E-09 |
-26.8218303 |
chr4_26789871_26790007 |
chr4 |
0 |
- |
9.76527E-06 |
2.75795E-05 |
-27.15229384 |
chr1_55370080_55370265 |
chr1 |
0 |
- |
1.33489E-60 |
3.89366E-58 |
-27.2302102 |
chr19_45619267_45619497 |
chr19 |
2 |
MARK4|PPP1R37 |
3.61011E-48 |
6.62101E-46 |
-27.29034787 |
chr16_33588844_33589010 |
chr16 |
1 |
ENPP7P13 |
8.03476E-26 |
4.0933E-24 |
-27.38040136 |
chr4_15907926_15908145 |
chr4 |
0 |
- |
2.8442E-50 |
5.76083E-48 |
-27.44818039 |
chr10_67155111_67155236 |
chr10 |
0 |
- |
4.83108E-11 |
4.17264E-10 |
-27.80333564 |
chr17_4278292_4278786 |
chr17 |
1 |
UBE2G1 |
9.00939E-24 |
3.82074E-22 |
-27.82640422 |
chr11_116344108_116344147 |
chr11 |
0 |
- |
0.000326633 |
0.000615342 |
-27.8388828 |
chr19_34245878_34246069 |
chr19 |
1 |
CHST8 |
6.69284E-06 |
1.96741E-05 |
-27.92649973 |
chr17_16864290_16864524 |
chr17 |
1 |
TNFRSF13B |
1.26001E-12 |
1.41214E-11 |
-28.2842299 |
chr11_82354426_82354705 |
chr11 |
2 |
MIR4300HG|RP11-179A16. |
6.09835E-07 |
2.30172E-06 |
-29.18358119 |
chr5_153676389_153676633 |
chr5 |
1 |
GALNT10 |
1.43406E-68 |
5.14737E-66 |
-29.70725527 |
chr10_134778418_134779164 |
chr10 |
3 |
LINC01166|LINC01167|LINC01168 |
5.1864E-178 |
1.2399E-174 |
-29.73911375 |
chr8_36995835_36995986 |
chr8 |
1 |
MIR1268A |
3.40507E-09 |
2.0846E-08 |
-29.75107115 |
chr2_213316451_213316721 |
chr2 |
1 |
ERBB4 |
3.25312E-05 |
8.05895E-05 |
-30.06050559 |
chr3_105601223_105601548 |
chr3 |
0 |
- |
3.99011E-26 |
2.08106E-24 |
-30.45696192 |
chr2_130693963_130694229 |
chr2 |
3 |
PLAC9P1|LINC01856|AC079776. |
1.8989E-114 |
1.6726E-111 |
-30.54653298 |
chr11_110986763_110986975 |
chr11 |
1 |
RP11-89C3. |
0 |
0 |
-30.61868687 |
chr4_184296182_184296485 |
chr4 |
2 |
RP11-451F20.|AC093844. |
3.2812E-82 |
1.64531E-79 |
-30.6235367 |
chr6_21452696_21452935 |
chr6 |
0 |
- |
4.66098E-09 |
2.77827E-08 |
-30.7652865 |
chr14_56669107_56669307 |
chr14 |
1 |
PELI2 |
0.005344129 |
0.006946025 |
-31.10639369 |
chr13_25141770_25142091 |
chr13 |
3 |
TPTE2P6|PSPC1P|RP11-556N21. |
2.11697E-25 |
1.04092E-23 |
-31.10709988 |
chr2_238796929_238797268 |
chr2 |
1 |
RAMP1 |
9.04001E-24 |
3.83251E-22 |
-31.35072812 |
chr17_6901611_6901751 |
chr17 |
4 |
ALOX12-AS1|RP11-589P10.|RP11-589P10.|ALOX12 |
0.00014541 |
0.000302279 |
-31.60823695 |
chr15_79299672_79299707 |
chr15 |
1 |
RASGRF1 |
0.004476538 |
0.005967375 |
-32.72230999 |
chr1_223353214_223353363 |
chr1 |
1 |
RP11-239E10 |
7.13359E-18 |
1.64468E-16 |
-33.00889209 |
chr1_113221602_113221746 |
chr1 |
2 |
CAPZA1|MOV10 |
8.44636E-34 |
7.62526E-32 |
-33.03827068 |
chr19_21860552_21861267 |
chr19 |
1 |
RP11-420K14. |
1.3469E-284 |
1.127E-280 |
-33.08420719 |
chr2_145505959_145506291 |
chr2 |
2 |
TEX41|AC023128. |
2.36911E-23 |
9.63501E-22 |
-33.13077409 |
chr21_29565178_29565285 |
chr21 |
1 |
LINC01695 |
0.000196292 |
0.000393653 |
-33.4915241 |
chr22_47442504_47442671 |
chr22 |
1 |
TBC1D22A |
2.44869E-31 |
1.87658E-29 |
-34.87001855 |
chr8_119449323_119449547 |
chr8 |
1 |
SAMD12 |
2.1869E-06 |
7.23312E-06 |
-36.3138769 |
chr3_6706652_6706860 |
chr3 |
2 |
AC069277.|GRM7-AS3 |
0.00028562 |
0.000547251 |
-39.63810451 |
chr8_129286309_129286584 |
chr8 |
0 |
- |
3.30084E-09 |
2.02608E-08 |
-39.75752508 |
chr12_64067778_64067878 |
chr12 |
3 |
MIR548Z|DPY19L2|RP11-415I12. |
0.000389652 |
0.000718873 |
-40.8170893 |
chr15_71187717_71187868 |
chr15 |
2 |
LRRC49|THAP10 |
0.004522921 |
0.006020138 |
-42.45762712 |
chr3_115099771_115100023 |
chr3 |
0 |
- |
0.002555418 |
0.003683188 |
-42.60471914 |
chr17_57187586_57187924 |
chr17 |
3 |
TRIM37|AC099850.|SKA2 |
2.39899E-60 |
6.93704E-58 |
-46.21784352 |
To identify the molecular pathways and functions potentially influenced by methylation changes in AN, we performed GO term and KEGG pathway enrichment analyses of the genes closest to the identified DMRs (within the gene body or within +/- 10kb of gene start/end sites) using the DAVID bioinformatics resources 6.8 (https://david.ncifcrf.gov/). When "Disease" was used for categorization, there were 5 charts categories (with a significant p value with Benjamini correction) of DMRs, the more significant charts being waist-hip ratio (n=9 counts, Benjamini 5.2e-3), tobacco use disorder (37 counts, Benjamini 2.8e-3), chemdependency (n=40 counts, Benjamini 1.2e-3), metabolic (n=49, Benjamini 3.1e-3) and neurological (32 counts, Benjamini 3.8e-3).
We next focused on the 153 DMRs, the String network analysis software identified a subnetwork, related to MAPK signaling pathway (hs04010, FDR 0.0026), PI3K-Akt signaling pathway (hsa04151, FDR 0.0232) and neurotrophin signaling pathway (hsa04550, FDR 0.0251)(https://string-db.org/cgi/network.pl?taskId=07ri9Goe1SzI) (Figure 1).
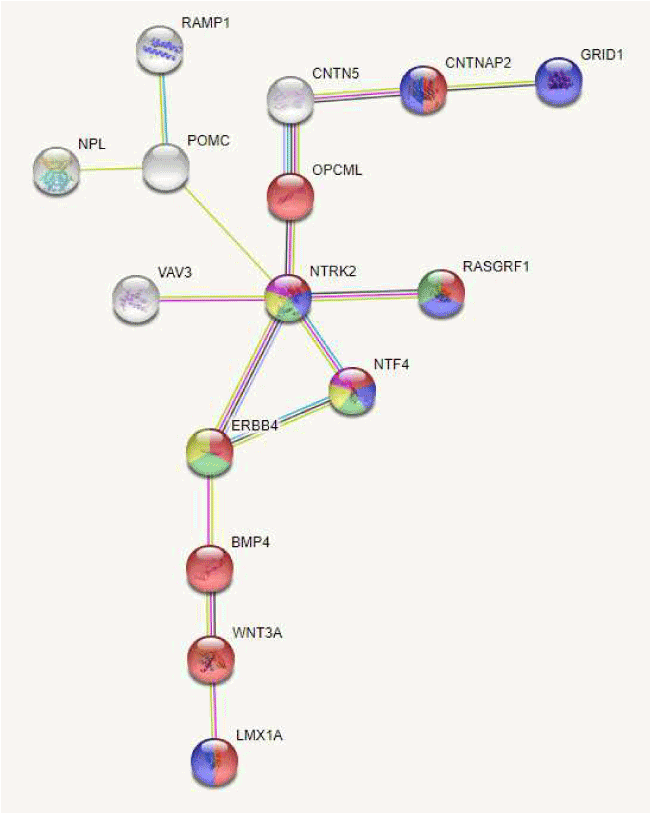
Figure 1. Protein subnetwork of the identified proteins constructed with the STRING software for enrichment analysis of proteins (PPI enrichment value 7.04e-08) showing a dysmethylation status (15 DMRs among the 119 identified DMRs including at least one coding or non-coding genes). Each node represents a protein entity. Gene Ontology identified functional enrichments for several biological process including neuron differentiation (GO:0030182, FDR 8.15e-06) and behavior (GO:0007610, FDR 0.00025)(in black), and KEGG pathways identified enrichments for several pathways including MAPK signaling pathway (hs04010, FDR 0.0026)(grey square), PI3K-Akt signaling pathway (hsa04151, FDR 0.0232)(white square) and Neurotrophin signaling pathway (hsa04550, FDR 0.0251) (grey triangle)(https://string-db.org/cgi/network.pl?taskId=07ri9Goe1SzI)
To the best of our knowledge, the present study presents the first genome-wide DNA methylation profiling of 6 male AN patients, using a high-throughput DNA methylation sequencing covering a large number of CpG sites on the human genome. Patients affected with AN restrictive type showed many differentially methylated sites, with significant between group differences corresponding to genes implicated in metabolic and nutritional status, psychiatric status and immune function. When we compared our DMCs list with the short list of Steiger and colleagues obtained in female AN patients, 32 genes were found in both lists [9]. Moreover, because the interaction between depression and anorexia nervosa was significant, we also identified six genes previously found in patients with major depressive disorder [8]. Taking into account that our approach was based on genome-wide methylation sequencing, we also identified additional candidate genes involved in psychiatric disorders (GRID1, NAALADL2-AS3, PDZD, CEP85L, GRIK3, SLC7A14), in metabolism regulation (lipids, ZFP36L1, CERK, ACSF3, EEPD1; glucose, KCNA7), and finally, in addiction (GDE1, CCKAR, PDYN). Moreover, we identified few genes previously associated with anorexia nervosa in genome-wide association studies (VGLL4, GRID1, WWOX, CAMK1D, SORCS2) (https://www.ebi.ac.uk/gwas/search?query=anorexia%20nervosa). Interestingly, the String network analysis software revealed a subnetwork related to neurotrophin signaling pathway (Figure 1). The reported data, in addition to the previous reported findings for BDNF, NTRK2 and NTRK3, point again neurotrophin signaling genes as key regulators of eating behavior.
To conclude, our data replicates several results concerning several target genes such as PRKAG2, RPTOR, and ICAM5 previously discussed [9], and identified novel signaling pathways involving PI3K-Akt and neurotrophin signaling pathway disturbed in anorexia nervosa. Future replication of findings in male AN patients will be a determinant.
Funding
This study was supported by Institut National de la Recherche Médicale (INSERM) and by Fondation Maladies Rares (program High throughput sequencing and rare disease, number #11632).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procédures performed in this study were in accordance with the ethical standards of our national research committee and with the 1964 Helsinki declaration and its later amendments. Informed written consent was obtained from all patients and parents included in the study.
We thank the families for their enthusiastic participation and all physicians from the different medical and psychiatric centers.
View supplementary data
- Bulik CM, Sullivan PF, Tozzi F, Furberg H, Lichtenstein P, et al. (2006) Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry 63: 305-312. [Crossref]
- Wang K, Zhang H, Bloss CS, Duvvuri V, Kaye W, Schork NJ, et al. (2011) A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry 16: 949-959. [Crossref]
- Boraska V, Franklin CS, Floyd JA, Thornton LM, Huckins LM, et al. (2014) A genome-wide association study of anorexia nervosa. Mol Psychiatry 19: 1085-1094. [Crossref]
- Duncan L, Yilmaz Z, Gaspar H, Walters R, GoldsteinJ, Anttila V, et al. (2017) Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry 174: 850-858. [Crossref]
- Huckins LM, Hatzikotoulas K, Southam L, Thornton LM, Steinberg J, Aguilera-McKay F, et al. (2018) Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry 23: 1181-1188. [Crossref]
- Thornton LM, Welch E, Munn-Chernoff MA, Lichtenstein P, Bulik CM (2016) Anorexia nervosa, major depression, and suicide attempts: shared genetic factors. Suicide Life Threat Behav 46: 525-534. [Crossref]
- Wade TD, Bulik CM, Neale M, Kendler KS (2000) Anorexia nervosa and major depression: shared genetic and environmental risk factors. Am J Psychiatry 157: 469-471. [Crossref]
- Shimada M, Otowa T, Miyagawa T, Umekage T, Kawamura Y, Bundo M, et al. (2018) Anepigenome-wide methylation study of healthy individuals with or without depressive symptoms. J Hum Genet 63: 319-326. [Crossref]
- Steiger H, Booij L, Kahan `, McGregor K, Thaler L, Fletcher E, et al. (2019) A longitudinal,epigenome-wide study of DNA methylation in anorexia nervosa: results in actively ill, partially weight-restored, long-term remitted and non-eating-disorderedwomen. J Psychiatry Neurosci 44: 1-9. [Crossref]
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. (2012) MethylKit: a comprehensive R package for the analysis of genome-wideDNA methylation profiles. Genome Biol 13: 80-87. [Crossref]

