Seawater-compensated fuel ballast systems maintain ship stability as fuel is spent but can introduce microorganisms that form biofilms, biodegrade fuel components, or enhance corrosion; all of which increase operating and maintenance costs. The aim of this study was to isolate planktonic bacteria from ballast tank fluids and taxonomically classify those that formed biofilm in culture. Twenty-two isolates were identified as belonging to seven genera based on 16S rRNA gene sequencing. Of the seven genera represented Alteromonas, Pseudoalteromonas, and Brevundimonas strains produced quantifiable biofilm in crystal violet assays. To test the hypothesis that the level of bulk nutrients would influence the extent of biofilm formation, isolates were grown in a conventional marine medium and marine medium supplemented with tryptone and yeast extract to represent standard and nutrient-replete media, respectively. While 80% of the Pseudoalteromonas strains produced 7 to 11-fold more biofilm in conventional medium vs. the supplemented medium, 64% of Alteromonas strains produced up to 50-fold less in the same medium. These results suggest that bulk nutrients influence the extent of biofilm formation in a taxa-specific fashion in these marine organisms. The sole Dasania marina isolate failed to display considerable biofilm growth in either media but was the only isolate to produce quorum sensing molecule(s), N-acyl homoserine lactone(s), in assays using an Agrobacterium tumefaciens reporter strain. Whether N-acyl homoserine lactones produced by D. marina could modulate biofilm formation in the other organisms isolated in this study would require further investigation.
marine biofilm, Dasania marina, ballast tank, 16S rRNA
Biofilms are a form of fouling where microorganisms attach to a surface and produce a protective matrix that allows them to persist in hostile environments [1]. Several factors have been shown to influence the extent of biofilm formation including nutrient availability [2,3] and quorum sensing [4]. Biofilms present in marine systems can lead to increased maintenance costs or material damage [5]. For example, in maritime transport biofilm that accumulates on the hull of a ship promotes macrofouling by barnacles and mussels, and the increased drag increases fuel consumption by an estimated 30 to 50% [6]. Other examples of increases in biofouling-related maintenance costs include an annual increase of 2 to 4% due to fouling of ship ballast water [7] and 20 to 30% due to corrosion [8].
The Navy uses a compensated fuel ballast system in several classes of its ships, including cruisers and destroyers. Each system consists of a series of tanks connected in sequence and as fuel is spent seawater gets pumped in to compensate for the lost volume [9]. While this mechanism maintains ship stability during deployment, there can be unintended consequences to the ballast system infrastructure or fuel when seawater microorganisms are introduced. Several studies have demonstrated that some microorganisms can metabolize hydrocarbons present in fuels or enhance corrosion of the system [10-12].
The conditions for planktonic growth can be found within a ballast system [13] and include water, a permissible growth temperature and carbon source, and salt and nutrients which vary depending upon the route of the ship and type of water used for compensation [14]. In 2001, Drake et al. estimated that the ballast water from a ship can harbor up to 109 planktonic cells per L [15], and although the ballast water gets ejected during refueling operations, biofilm material remains attached to the walls of the system [16]. High-throughput DNA sequencing studies have shown that the composition and relative abundance of dominant taxa in bulk seawater differ from that within biofilms that form on surfaces submerged in seawater [17,18] suggesting that certain taxa, not all, contribute to biofilm formation. The aim of this study was to isolate planktonic bacteria from ballast tank fluids, classify those that formed biofilm in culture, and determine if the level of bulk nutrients in growth media influenced the amount of biofilm produced.
Bacterial strains and growth conditions
Bacterial isolates were obtained from the ballast tank fluids of four US Navy ships in 2014: USS Sampson (seawater in tank for 8 months), USS Higgins (seawater in tank for 4-6 months), USS Bunker Hill (seawater in tank 4 months), and USS Lake Champlain (seawater in tank 1 week). Pooled ballast tank water was plated onto Marine Agar 2216 (BD Difco) within 48 hours of receipt and plates were incubated at 26°C. Isolated colonies were re-streaked and grown for a minimum of three transfers to obtain pure cultures. Freezer stocks were made in Marine Broth 2216 (BD Difco) (MB), a common medium for culturing heterotrophic marine organisms, supplemented with 20% glycerol and stored at – 80°C. Culturing of strains was carried out in MB and a nutrient-replete Marine Broth (MB/TY), which was MB supplemented with 5 g of tryptone and 4 g of yeast extract per L.
The reporter strain used for the detection of N-acyl homoserine lactones (AHL), Agrobacterium tumefaciens KYC55 (pJZ372, pJZ384, pJZ410) [19], was grown in Luria-Bertani broth (LB) or AT medium supplemented with spectinomycin (100 µg/mL) gentamycin (20 µg/mL) and tetracycline (2 µg/mL) [20]. This reporter strain detects a wide range of exogenous AHL of different lengths and compositions including 3-oxo-C4- to 3-oxo-C16-HSL and C4- to C18-HSL. The AHL-positive control strain, Sinorhizobium meliloti RM41, was grown in LB supplemented with 2.5 mM of CaCl2 and 2.5 mM of MgSO4.
Taxonomic classification of ballast tank isolates
To identify bacterial isolates, genomic DNA was extracted using the PureLink Microbiome DNA Purification Kit (ThermoFisherScientific, US) following the manufacturer’s instructions. The 16S rRNA gene was amplified by PCR using the forward primer (fd1) 5’AGAGTTTGATCCTGGCTCAG3’ and reverse primer (rp2) 5’ACGGCTACCTTGTTACGACTT3’ set [21] and DreamTaq PCR Master Mix (ThermoFisherScientific). The PCR reactions were sequenced at the sequencing facility at the University of Texas at Austin using the forward and reverse primers listed above and 5’CAGCAGCCGCGGTAA3’ (519F) [22]. The Basic Local Alignment Search Tool (BLASTN) was used to align and identify each sequence. Sequences were submitted to GenBank under the accession numbers MK250492-MK250514. The sequence identifications were verified using the Ribosome Database Project Naive Bayesian rRNA Classifier Version 2.11 [23].
Assays for biofilm production and planktonic growth
Strains were grown overnight on Marine Agar and individual colonies were inoculated into 4 mL MB or MB/TY and incubated at 26°C with shaking (100 rpm) for 2 d. The starter cultures were observed for the presence of ring and floating biofilms at 2 d and 7 d, respectively and were used to inoculate 96-well plates for the crystal violet assay.
Crystal violet assays were performed as described [24]. Briefly, starter cultures were seeded into 96-well round-bottom plates at 1:100 dilution (OD600 nm=1.0). The plates were incubated at 26°C for 2 d with shaking (100 rpm). The liquid culture was transferred to a 96-well flat-bottom plate to measure planktonic growth at OD600 using a Synergy HT microplate reader (Biotek Instruments, US), and the biofilm that remained attached to the wells of the plate was washed, stained with 0.1% crystal violet, and quantified by measuring the OD550 as described by O’Toole [24].
AHL detection
A modified version of the procedure by Joelsson [20] was used to recover AHL from the supernatant of bacterial cultures. Briefly, 4 mL of log-phase cultures grown in MB were centrifuged at 13,500 x g for 3 min and the supernatant was subjected to extraction with 4 mL of ethyl acetate. The residue was resuspended in 400 μL (10X extract) acetonitrile and stored in a glass vial at -20 oC.
A variation of the method by Cha [25] was used to detect AHL on solid media. Briefly, the AHL-reporter strain A. tumefaciens KYC55 was grown on LB agar with antibiotics at 26 oC for 2 d, and isolated colonies were inoculated into 0.6 mL AT medium with antibiotics and grown for 24 h. The cells were seeded into 20 mL of warm melted AT agar (0.8%) supplemented with antibiotics and 40 µL of 20 mg/mL X-gal and poured to make AT agar plates. Five µL aliquots from each 10X extract were spotted onto AT agar plates, and plates were incubated at 26°C for 24 h. To quantify AHL in broth cultures, β-galactosidase assays were conducted as described by Joelsson [20]. The 10X extract from S. meliloti RM41 and 100 µM of the synthetic C8-HSL were included as positive controls.
Identification of ballast tank isolates
Twenty-two bacterial isolates were classified into three dominant phyla, with all but two belonging to Gammaproteobacteria. SDH6 was classified as Brevundimonas mediterranea belonging to Alphaproteobacteria and SD20 was classified as Bacillus adhaerens belonging to the Firmicutes (Table 1). SD20 was identified as the only gram-positive bacterium among the cohort. Of the 21 gram-negative strains, eight of the Gammaproteobacteria were classified as Alteromonas oceani S35 and three as A. tagae BCRC 17571. Five isolates were assigned to the genus Pseudoalteromonas, two were identified as Halomonas axialensis and the remaining Gammaproteobacteria were single organisms from the genera Dasania and Marinobacter. These data are consistent with a recent high throughput sequencing study showing that the most abundant Gammaproteobacterial OTUs from ballast waters from five separate ships belonged to Alteromonadales, which encompass the Alteromonas and Pseudoalteromonas genera [26].
Table 1. Identification of bacterial isolates based on 16S rRNA gene classification
Isolate |
Strain ID (GenBank closest match) |
Maximum identity (%) |
Accession No. |
Gammaproteobacteria |
SD3 |
Alteromonas oceani S35 |
99.89 |
NR 159349.1 |
SD4 |
Alteromonas oceani S35 |
98.84 |
NR 159349.1 |
SD8 |
Alteromonas oceani S35 |
98.85 |
NR 159349.1 |
SD9 |
Alteromonas oceani S35 |
99.88 |
NR 159349.1 |
SD13 |
Alteromonas oceani S35 |
99.66 |
NR 159349.1 |
SD17 |
Alteromonas oceani S35 |
98.28 |
NR 159349.1 |
SD18 |
Alteromonas oceani S35 |
98.41 |
NR 159349.1 |
SD32 |
Alteromonas oceani S35 |
99.78 |
NR 159349.1 |
SD5 |
Alteromonas tagae BCRC 17571 |
98.93 |
NR 043977.2 |
SD7 |
Alteromonas tagae BCRC 17571 |
99.26 |
NR 043977.2 |
SD16 |
Alteromonas tagae BCRC 17571 |
100.00 |
NR 043977.2 |
|
|
|
|
SD1 |
Pseudoalteromonas shioyasakiensis SE3 |
99.57 |
NR 125458.1 |
SD10 |
Pseudoalteromonas shioyasakiensis SE3 |
99.57 |
NR 125458.1 |
SD31 |
Pseudoalteromonas undina NBRC 103039 |
99.89 |
NR 114191.1 |
SD1A |
Pseudoalteromonas tetraodonis GFC IAM 14160 |
100.00 |
NR 041787.1 |
SD1B |
Pseudoalteromonas tetraodonis GFC IAM 14160 |
100.00 |
NR 041787.1 |
|
|
|
|
SD2D |
Halomonas axialensis Althf1 |
99.93 |
NR 026219.1 |
SD2E |
Halomonas axialensis Althf1 |
99.86 |
NR 026219.1 |
|
|
|
|
SD1D |
Dasania marina DMS 21967 KOPRI 20902 |
98.50 |
NR 043175.1 |
|
|
|
|
SDH2 |
Marinobacter adhaerens HP15 |
99.93 |
NR 037108.1 |
|
|
|
|
Alphaproteobacteria |
SDH6 |
Brevundimonas mediterranea V4.BO.10 |
99.85 |
NR 037108.1 |
|
|
|
|
Firmicutes |
SD20 |
Bacillus aquimaris TF-12 |
99.20 |
NR 025241.1 |
Biofilm production in MB and MB/TY
All isolates were initially screened for biofilm production by observing cultures grown in glass tubes (Figure 1). In doing so, two biofilm morphologies were observed: a ring and a floating disk, described by Mosharaf [27] as solid-air-liquid (SAL) and air-liquid (AL) biofilms, respectively. The ring morphology adhered to the wall of the glass tube and formed at the interface between the air and liquid culture (Figure 1, panel A). The floating disk morphology was attached to the surface of the culture and may or may not appear to adhere to the tube (Figure 1, panel B). All isolates except for the Marinobacter strain formed at least one of the two types of biofilm observed (Table S1). Alteromonas strains produced thick ring and floating biofilms with the exception of SD16, one of the three A. tagae isolates. It did not produce a visible ring and produced only a weakly visible floating biofilm. The five Pseudoalteromonas strains also produced both morphologies, but to a lesser extent than the Alteromonas strains. Finally, the single strains of Dasania, Marinobacter, and Bacillus produced very little or no detectable biofilms in glass tubes (Table S1).
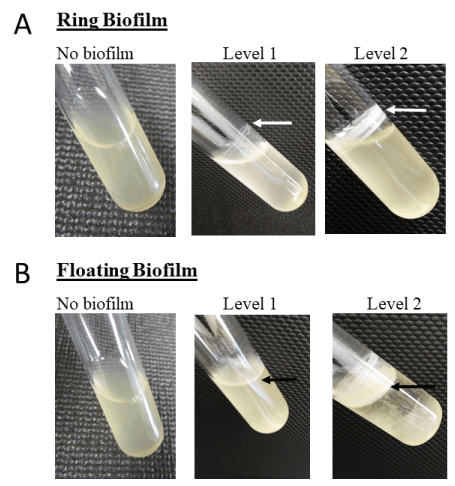
Figure 1. Images of biofilm formed in glass tubes. panel A, isolates were grown in MB or MB/TY for 2 d at 26 °C with agitation (100 rpm), and biofilms were observed and classified as level 1 (weak/thin biofilm) or level 2 (thick biofilm). panel B, strains were grown as described but were incubated further under static conditions for 7 d at 19 °C. Arrows point to biofilm
Sixteen of the 22 isolates produced quantifiable biofilm in crystal violet assays in either MB or MB/TY, with OD550 values greater than the media-only control (OD550=0.04) (Figure 2). This included all Alteromonas strains (Figure 2, panel A), four of the five Pseudoalteromonas strains (Figure 2, panel B), and the sole Brevundimonas strain (Figure 2, panel C). The remaining six strains produced very little to no biofilm when grown in either media and included strains belonging to the genera Halomonas, Dasania, Marinobacter, and Bacillus.
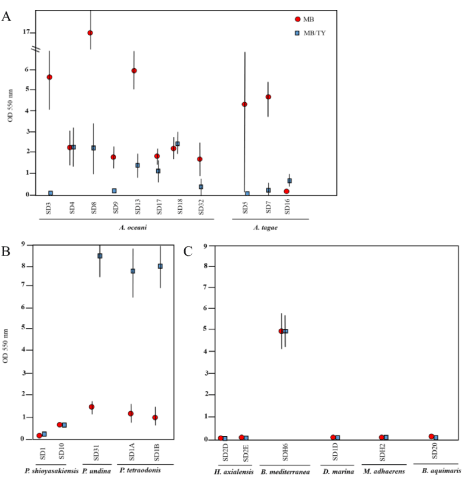
Figure 2. Biofilm quantification (OD 550 nm) in microtiter plates. Isolates were inoculated 1:100 (at an OD600 = 1) into microtiter plates, cultured for 2 d, and the biofilm material was stained as described in materials and methods. The OD550 for each strain was determined by subtracting the OD550 of the media-only wells. Data shown are representative of two independent experiments performed on different days. The results are the mean ± standard deviation (n = 8). Standard deviations below 0.2 are not shown
Although there were eight A. oceani strains and all produced quantifiable biofilm material, there was wide variation in their abilities to do so (Figure 2). SD8 produced significantly more biofilm material than any of the other strains when grown in MB (OD550=16.95±1.91). All but three isolates (SD4, SD17, SD18) produced more biofilm material when grown in MB vs MB/TY (Figure 2, panel A), with SD3 producing over 50-fold more. There was also variation in biofilm formation in the A. tagae isolates in that SD7 produced considerably more biofilm in MB than in MB/TY, and SD16, a weak biofilm producer, produced more biofilm in MB/TY. Interestingly, A. tagae (SD5), similar to SD7, produced more biofilm in MB (OD550 > 4), but the biofilm material was not consistently stable to withstand the washes and resulted in some replicates being weakly stained in the same experiment. This accounted for the high standard deviation observed for SD5 grown in MB media. Furthermore, quantification of crystal violet in nutrient-replete medium was difficult since this strain produced a gel-like “pellicle” akin to the Alphaproteobacteria member Gluconacetobacter xylinus [28], which was ejected during the wash steps. Floating biofilms have been extensively studied in gram-positive bacteria such as Bacillus thuringiensis [29] or B. subtillus [28], where components of the matrix which allow it to float consist of proteins that form amyloid-like fibers and a hydrophobic layer. It is unknown if the gel-like matrix formed by SD5 in nutrient-replete marine medium consisted of similar proteins. Nevertheless, it was one of the only isolates to form this type of floating biofilm in microtiter plates.
In contrast to the Alteromonas strains which generally produced more biofilm in MB than in MB/TY, three of the five Pseudoalteromonas strains produced 7- to 11-fold more biofilm in MB/TY (Figure 2). This included the sole P. undina strain (SD31) and the two P. tetraodonis strains (SD1A and SD1B). Of the two P. shioyasakiensis strains (SD1 and SD10), only one produced quantifiable biofilm and did so at equivalent levels in both growth media. That the majority of the Alteromonas isolates produced more robust biofilms in MB, while isolates belonging to Pseudoalteromonas did so in MB/TY, supports the hypothesis that the level of bulk nutrients influence the extent of biofilm formation, and that it did so in a taxa-specific fashion.
Finally, the sole B. mediterranea strain produced comparable levels of biofilm in both growth media (OD550=4.74±0.78 and 4.72±0.69) but the lowest overall planktonic growth (OD600) of all strains in MB (Table S2). The remaining five strains produced levels of biofilm that were comparable to background (OD550 ≤ 0.04). For all isolates except SD5 (an Alteromonas strain), planktonic growth in the same cultures was higher in MB/TY than MB (Table S2).
AHL detection
Since quorum sensing is a variable that can influence biofilm formation and all of the biofilm-forming isolates in this study were gram-negative, they were subsequently screened on plates for the ability to produce AHL using the reporter strain A. tumefaciens KYC55. As shown in Figure 3, only D. marina (SD1D) produced AHL detectable by this reporter and did so in both growth media. Figure 3 panel C shows the controls used in this assay, demonstrating that the synthetic C-8 AHL and AHL produced by S. meliloti RM41 were detectable. When further experiments were performed to quantify β-galactosidase activity, the levels of activity of SD1D grown in either MB or MB/TY were comparable to levels from an extract of the positive control strain, S. meliloti RM41, and the synthetic C8-HSL (Figure 4). Also shown in the figure are extracts from SDH6 (B. mediterranea) grown in MB and MB/TY. This strain produced comparable biofilm in both types of media, however AHL were not detectable. Extracts from the other isolates were negative for AHL production by the reporter strain (data not shown). This negative result for the Pseudoalteromonas strains was not expected as extracts from P. shioyasakiensis and P. tetraodonis strains isolated from sediments in the Zhejiang Province demonstrated a positive response in reporter assays using a different A. tumefaciens A136 reporter strain that responds to C6- to C14-HSL [30]. This range is within that of the reporter strain used in this study. That some of the isolates produced AHL that were not detected by the reporter cannot be ruled out but was beyond the scope of this study. Indeed, Huang et al. concluded that AHL bioassay results are not always consistent among different screening methods [31]. However, only the D. marina isolate (SD1D), which failed to produce quantifiable biofilm under both nutrient conditions, tested positive for AHL using the reporter. Whether or not AHL released by SD1D could have an impact on biofilm formation when co-cultured with the other isolates is currently being investigated.
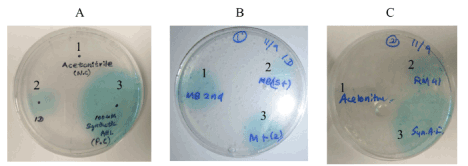
Figure 3. AHL screen using a reporter strain. The reporter strain A. tumefaciens KYC55 was seeded into warm AT medium containing antibiotics and X-gal as described in the materials and methods. 10X extracts from bacterial isolates were spotted onto AT agar plates. The turquoise zone is indicative of the presence of AHL. panel A, AHL production of SD1D grown in MB. 1 is acetonitrile (negative control), 2 is 10X extract from SD1D, 3 is synthetic C8-HSL (100 µM solution). panel B, AHL production of SD1D in MB (1) or MB/TY (2 and 3). panel C, AHL production in positive and negative controls. 1 is acetonitrile (negative control), 2 is 10X extract from S. meliloti RM41 (positive control extract), and 3 is synthetic C8-HSL (positive control)
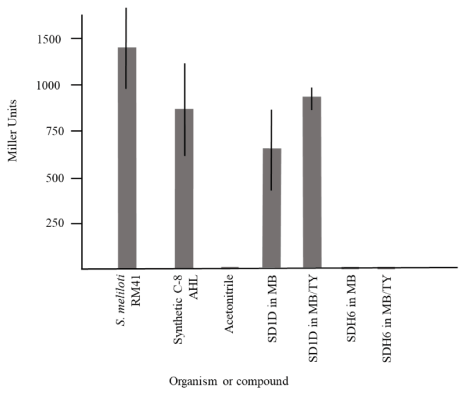
Figure 4. AHL quantification using β-galactosidase assays. One hundred microliters of the reporter strain A. tumefaciens KYC55 were seeded into 2 mL of AT medium. Ten microliters of C8-HSL (positive control) or 10X extracts were added, the culture was grown to an OD600 of 0.2 to 1, and β-galactosidase assays were conducted as described [20]. Data shown are the mean ± standard deviation (n = 2, two independent experiments). Standard deviations below 3 are not shown
Approximately 70% (16 of 22) of isolates recovered from ballast tanks produced quantifiable biofilms in at least one growth media tested. Alteromonas isolates displayed enhanced planktonic growth in nutrient-replete medium, however biofilm formation was reduced under these same conditions for 70% (8 of 11) of them, suggesting that increased bulk nutrients limit biofilm formation for this group. Indeed, studies in other biofilm-producing strains, including Lysteria monocytogenes, showed a similar trend [32]. Alternatively, and unexpectedly, Pseudoalteromonads produced more biofilm when bulk nutrients were in excess.
A recent study demonstrated that Pseudoalteromonas strains were both cultivable on hydrocarbon-enriched media and abundant in oiled mesocosms [33], consistent with Pseudoalteromonas strains being abundant in hydrocarbon-laden environments, such as ballast tanks that hold fuel. Furthermore, Pseudoalteromonas, Alteromonas, Halomonas, and Marinobacter strains were identified as hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill in 2010 [34]. Whether or not these ballast fluid isolates can biodegrade fuel was beyond the scope of this study but is currently being investigated.
The reporter strain A. tumefaciens was selected for the screen since it detects a wide range of exogenous AHL [19] and those associated with biofouling include C4- to C12-HSL and 3-oxo-C6- to 3-oxo-C12-HSL [35]. AHL were not detected in culture extracts from the biofilm-forming isolates in this study, suggesting that strains recovered from ballast tanks may not use AHL for biofilm formation, use AHL that is out of the range of the reporter strain, or possibly use a different quorum sensing system altogether if at all. Whether AHL produced by the D. marina isolate could modulate biofilm formation is being investigated. To the authors knowledge, very little is known about D. marina, outside of its identification and characterization in 2007 [36]. Thus, the possibility of this organism using its quorum sensing system has yet to be explored and is of interest for future studies.
The authors would like to thank the Alternate Fuels Multidisciplinary University Research Initiative at the University of Oklahoma for procuring the ballast tank water; Kenneth Sandoval for assistance in obtaining pure cultures of isolates; JeanKathleen Cala and Jeanette Cala for technical assistance; and Drs. Juan Gonzalez and Jun (Jay) Zhu at the University of Texas at Dallas and the Perelman School of Medicine, respectively for providing control strains and protocols.
The authors declare that they have no competing interests.
This work was supported by startup funds for Athenia Oldham.
- Lee KW, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, et al. (2014) Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J 8: 894-907. [Crossref]
- Ryu J, Kim H, Beuchat LR (2004) Attachment and biofilm formation by Escherichia coli 0157:H7 on stainless steel as influenced by exopolysaccharide production, nutrient availability, and temperature. J Food Prot 67: 2123-2131. [Crossref]
- Cherifi T, Jacques M, Quessy S, Fravalo P (2017) Impact of nutrient restriction on the structure of Listeria monocytogenes biofilm grown in a microfluidic system. Front Microbiol 8: 864. [Crossref]
- Sadekuzzaman M, Yang S, Mizan MFR, Ha SD (2015) Current and recent advanced strategies for combating biofilms. Compr Rev Food Sci 14: 491-509.
- Carvalho CCCRD (2018) Marine biofilms: A successful microbial strategy with economic implications. Front Mar Sci 5: 1-11.
- Shultz MP, Bendick JA, Holm ER, Hertel WM (2011) Economic impact of biofouling on a naval surface ship. Biofouling 27: 87-98. [Crossref]
- Fernandes JA, Santos L, Vance T, Fileman T, Smith D, et al. (2016) Costs and benefits to European shipping of ballast-water and hull fouling treatment: Impacts of native and non-indigenous species. Marine Policy 64: 148-155.
- Skovhus TL, Enning D, Lee JS (2017) Microbiologically-influenced corrosion in an upstream oil and gas industry, Boca Raton, FL: CRC Press.
- US Environmental protection agency. Appendix A, compensated fuel ballast: Nature of discharge for the “phase I final rule and technical development document of uniform national discharge standards (UNDS)” 1999. Reference number EPA-842-R-99-001. (https://www.epa.gov/vessels-marinas-and-ports/uniform-national-discharge-standards-unds-phase-i-final-rule) accessed 12.09.2019.
- Aktas DF, Lee JS, Little BJ, Duncan KE, Perez-Ibarra BM (2013) Effects of oxygen on biodegradation of fuels in a corroding environment. Int Biodeterior & Biodegradation 81: 114-126.
- Liang R, Aktas DF, Aydin E, Bonifay V, Sunner J, et al. (2016) Anaerobic biodegradation of alternative fuels and associated biocorrosion of carbon steel in marine environments. Environ Sci and Technol 50: 4844-4853.
- Liang R, Aydin E, Le Borgne S, Sunner J, Duncan KE, et al. (2018) Anaerobic biodegradation of biofuels and their impact on the corrosion of a Cu-Ni alloy in marine environments. Chemosphere 195: 426-436.
- Pickard GL, Emery WJ (1982) Descriptive physical oceanography: An introduction, fourth ed. Pergamon Press, Oxford Wiley. New York.
- Heyer A, D’Souza F, Morales CFL, Ferrari G, Mol JMC, et al. (2013) Ship ballast tanks: a review from microbial corrosion and electrochemical point of view. Ocean Eng 70: 188-200.
- Drake LA, Choi KH, Ruiz GM, Dobbs FC (2001) Global redistribution of bacterioplankton and virioplankton communities. Biol Invasions 3: 193-199.
- Baier RE, Forsberg RL, Meyer AE, Lundquist DC (2014) Ballast tank biofilms resist water exchange but distribute dominant species. Manag Biol Invasions 5: 241-244.
- Oldham AL, Steinberg MK, Duncan KE, Makama Z, Beech I (2017) Molecular methods resolve the bacterial composition of natural marine biofilms on galvanically coupled stainless-steel cathodes. J Ind Microbiol and Biotechnol 44: 167-180.
- Dang H, Lovell CR (2016) Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80: 91-138. [Crossref]
- Zhu J, Chai Y, Zhong Z, Li S, Winans SC (2013) Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69: 6949-6953. [Crossref]
- Joelsson AC, Zhu J (2006) LacZ-based detection of acyl-homoserince lactone quorum-sensing signals. Curr Protoc Microbiol. [Crossref]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697-703. [Crossref]
- Ovreas L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63: 3367-3373. [Crossref]
- Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261-5267.
- O'Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp 47: 2437. [Crossref]
- Cha C, Gao P, Chen YC, Shaw PD, Farrand SK (1998) Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. MPMI 11: 1119-1129. [Crossref]
- Brinkmeyer R (2016) Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing. Pollution Bull 107: 267-285. [Crossref]
- Mosharaf MK, Tanvir MZH, Haque MM, Haque MA, Khan MAA, et al. (2018) Metal-Adapted bacteria isolated from wastewaters produce biofilms by expressing proteinaceous curli fimbriae and cellulose nanofibers. Front Microbiol 8: 1334. [Crossref]
- Armitano J, Méjean V, Jourlin-Castelli C (2014) Gram-negative bacteria can also form pellicles. Environ Microbiol Rep 6: 534-544. [Crossref]
- El-Khoury N, Majed R, Perchat S, Kallassy M, Lereclus D, et al. (2016) Spatio-temporal evolution of sporulation in Bacillus thuringiensis biofilm. Front Microbiol 7: 1222. [Crossref]
- Yu Z, Ding Y, Yin J, Yu D, Zhang J, et al. (2018) Dissemination of genetic acquisition/loss provides a variety of quorum sensing regulatory properties in Pseudoalteromonas. Int J Mol Sci 19: 3636. [Crossref]
- Huang YL, Ki JS, Case RJ, Qian PY (2008) Diversity and acyl-homoserine lactone production among subtidal biofilm-forming bacteria. Aquat Microb Ecol 52: 185-193.
- Kadam SR, Den Besten HM, Van Der Veen S, Zwietering MH, Moezelaar R, et al. (2013) Diversity assessment of Listeria monocytogenes biofilm formation: impact of growth condition, serotype, and strain origin. Int J Food Microbiol 165: 259-264. [Crossref]
- Chronopoulou PM, Sanni GO, Silas-Olu DI, van der Meer JR, Timmis KN, et al. (2015) Generalist hydrocarbon-degrading bacterial communities in the oil-polluted water column of the North Sea. Microb Biotechnol 8: 434-447. [Crossref]
- Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, et al. (2013) Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J 7: 2091-2104. [Crossref]
- Liu J, Fu K, Wang Y, Wu C, Li F, et al. (2017) Detection of diverse N-acyl-homoserine lactones in Vibrio alginolyticus and regulation of biofilm formation by N-(3-oxodecanoyl) homoserine lactone in vitro. Front Microbiol 8: 1097. [Crossref]
- Lee YK, Hong SG, Cho HH, Cho KH, Lee HK (2007) Dasania marina gen. nov., sp. nov., of the order Pseudomonadales, isolated from Arctic marine sediment. J Microbiol 45: 505-509. [Crossref]




