Abstract
Idiopathic pulmonary fibrosis (IPF) is the most severe lung fibrotic form and very few pharmacological therapies are available at present. Key events in the onset of the disease are the activation of fibroblasts to myofibroblasts and the production and release of extracellular matrix (ECM) and molecular factors.
Primary murine lung fibroblasts were isolated and their activation induced by Bleomycin (BLM) treatment. Extracellular Vesicles (EV) were isolated and protein extracted. Released soluble proteins (Secretome) and EV-derived proteins were reduced, alkylated and trypsin digested. A nano-LC-MS/MS SWATHTM approach was used for the proteomics analyses.
Specific proteins with a putative role in the transition from physiological to fibrotic conditions, such as several matrix metalloproteinases (MMPs), osteopontin (OPN), chitinase-3-like protein1 (CHI3L1) and CD44 resulted differentially released from BLM-treated fibroblasts as compared with untreated lung fibroblasts. Our results provide further understanding of the pathophysiological features of lung fibrosis, and suggest specific target for pharmacological treatments.
Key words
bleomycin, idiopathic pulmonary fibrosis, lung fibroblasts, proteomics analysis
Abbreviations
BCA: Bicinchoninic Acid; BLM: Bleomycin; CHI3L1: Chitinase-3-Like Protein 1; DAB: Dark Brown Stain; DMEM-HG: Dulbecco’s Modified Eagle Medium High Glucose, 4500 mg/L; ECM: Extracellular Matrix; EMT: Epithelial-Mesenchymal Transformation; FBS: Fetal Bovine Serum; FSP-1: Fibroblast-Specific Protein 1; GSN: Gelsolin; H&E: Haematoxylin and Eosin; H2O2: Hydrogen Peroxide; IIPs: Idiopathic Interstitial Pneumonias; IPF: Idiopathic Pulmonary Fibrosis; MANOVA: Multifactor Analysis Of Variance; MMP-1: Matrix Metalloproteinases-1; MMP-2: Matrix Metalloproteinases-2; MMP-3: Matrix Metalloproteinases-3; MMP-7: Matrix Metalloproteinases-7; MMP-9: Matrix Metalloproteinases-9; MMPs: Matrix Metalloproteinases; OPN: Osteopontin; PBS: Phosphate-Buffered Saline; PDGF: Platelet-Derived Growth Factor; PRDX2: Peroxiredoxin 2; PRDX6: Peroxiredoxin 6; RNS: Reactive Nitrogen Species; ROS: Reactive Oxygen Species; S100A4: S100 Calcium-Binding Protein A4; Tagln-2: Transgelin-2; TGF-β1: Transforming Growth Factor-β1; TNF-α: Tumor Necrosis Factor-α; Vim: Vimentin; α -sma: α-Smooth Muscle Actin
Introduction
Fibrotic lung diseases represent a broad spectrum of pathologies characterized by different degrees of inflammation and fibrosis. In this group of disorders, the highest degree of fibrosis is expressed in idiopathic pulmonary fibrosis (IPF), whose pathogenesis is still unclear, thus its prognosis is highly unfavorable [1]. Specific therapeutic treatments are still missing, anti-fibrotic therapy with Pirfenidone or the tyrosine kinase inhibitor Nintendanib has been demonstrated only to reduce disease progression [2]. During pathology onset, repetitive damage events lead epithelial cells to apoptosis, fibroblasts form “foci” and ECM components accumulate. Fibroblasts activate to myofibroblasts and express several cytokines and growth factors, such as PDGF, TGF-β1 and TNF-α, which stimulate cell migration and proliferation and extracellular matrix accumulation. Myofibroblasts are the major determinants of connective tissue remodeling in fibrotic disorders being also responsible for excessive ECM deposition, alveolar epithelial cell apoptosis and progressive destruction of lung parenchyma. All these processes end up to the progressive functional impairment of the lungs and, ultimately, to respiratory failure. Bleomycin (BLM) is the most widely used drug to induce pulmonary fibrosis in animals, being able to reproduce at least the main histologic characteristics observed in patients with IPF [3-5]. Presently, despite a better comprehension of fibrotic lung diseases lung transplantation remains the only real therapeutical opportunity for advanced IPF patients [6-8]. In this setting, we evaluated the modulation of protein synthesis and release in the BLM-induced phenotype switch from fibroblasts to myofibroblasts. In brief, secretome and proteins of microvesicles released by BLM-treated lung fibroblasts were analyzed to obtain a general overview of the proteins released from activated cells in order to better understand the pathophysiological features of lung fibrosis.
Methods
Lung fibroblast isolation and culture
Lungs of healthy mice (8-10 weeks old) were explanted and tissue cut into small pieces (2-4 mm2). Wild-type C57BL/6 female and male mice (8-10 weeks old) were purchased from Harlan Laboratories. National guidelines for the care and the use of research animals, notified by the Italian ministry of health in accordance with Italian law (D.L. 116/92, implementation of EEC directive 609/86) were followed. The study was conducted in collaboration with the Fondazione Toscana Gabriele Monasterio and the Center of Experimental Biomedicine, CNR, in Pisa.
Tissue fragments were digested using 1 mg/ml Clostridial collagenase (Sigma-Aldrich, Milan, Italy) in phosphate-buffered saline (PBS) incubating at 37°C for 1 hour. After the enzymatic digestion, tissue fragments were centrifuged at 300 x g for 10 minutes at room temperature, resuspended in cell growth medium (Dulbecco’s Modified Eagle Medium High Glucose, 4500 mg/L (DMEM-HG), supplemented with 10% FBS, 2 mM L-glutamine and 1X penicillin-streptomycin solution) and cultured under standard conditions at 37°C for at least 24 hours. The experiments were conducted with cells at the third passage.
MTT cell proliferation assay
For MTT Cell Proliferation Assay, lung fibroblasts were plated in 96-well dishes (2000 cells/well). Three replicates for each experimental point were prepared. Cells were treated for 24, 48, 72 hours with the following concentrations of BLM: 0.5 μg/ml; 1.5 μg/ml; 2.5 μg/ml; 3.5 μg/ml. 20 μl of MTT Reagent (CellTiter 96® Assay, Promega) were added for each well and cells were incubated up to 4 hours to 37°C, until purple precipitate was visible. At the end, the absorbance at 490 nm was recorded.
Secretome collection
Approximately 106 cells were seeded into 100 mm diameter culture dishes and cultured until cell density reached 90% confluence. For secretome analysis 3 dishes/experimental point (BLM-treated cells and control cells) containing 7 mL of culture FBS free-medium without phenol red were prepared. Conditioned medium was collected and centrifuged at 300 x g to eliminate cell debris. Salts were removed and protein concentrated by using Amicon Ultra-3 (Millipore) following the manufacturer’s recommendations.
Extracellular vesicles (EV) isolation
For EV isolation, 20 dishes/experimental point (BLM-treated cells and control cells), each containing 7 mL of culture medium, were prepared and cells were cultured with 100 000 x g ultracentrifuged FBS to avoid bovine vesicle contaminations. Vesicles were isolated through differential centrifugations using the protocol described by Comelli et al. (2014) with minor changes [9]. Briefly, medium was centrifuged at 2000 x g for 30 minutes to discard apoptotic blebs and at 110 000 x g for 1 h at 4°C to obtain microvesicles and exosomes (EV). The pellet was washed with PBS centrifuging for 2 hours at 110 000 x g at 4°C and the resulted pellet stored at -80°C until analysis.
In order to extract protein for MS analyses, vesicle pellets were lysed through 5 frozen-thaw cycles: 30s in liquid nitrogen and then thawed at 50°C for 2 min; finally the samples were sonicated for 5 min.
Sample preparation for MS analysis
For secretome and EV samples, protein concentration was determined by bicinchoninic acid (BCA assay, Thermo Fisher). 40 µg of each sample were processed, reducted with 5 mM DTT at 80°C for 30 min, then cooled to room temperature and alkylated for 30 min at 37°C with addition of iodoacetamide up to 10 mM final concentration. Digestion was performed by adding 0.50 µg of trypsin sequencing grade (Roche) at 37°C overnight and peptide mixtures were acidified with 10-20 µl of 1% formic acid.
Nano-LC-MS/MS SWATHTM analysis
Peptides were chromatographically separated using a nano-HPLC system (Eksigent, ABSciex). A first load step pre-concentrated the sample in a pre-column cartridge (PepMap-100 C18 5 µm 100 A, 0.1 x 20 mm, Thermo Scientific, USA) and then peptides were separated in a C18 PepMap-100 column (3 µm, 75 µm x 250 mm, Thermo Scientific, USA) at a flow rate of 300 nL min-1. Runs were performed with eluent A (Ultrapure water, 0.1% FA) under 70 min linear gradient from 5 to 40% of eluent B (ACN/0.1% FA) followed by 10 min of a purge step and 20 min re-equilibration step. Peptides eluted from chromatography were directly processed using TripleTOFTM 5600 mass spectrometer (ABSciex, USA) equipped with a DuoSprayTM ion source (ABSciex, USA). Data were acquired using the new SWATHTM method for shotgun data independent MRM quantification. For library, MS/MS data were processed with ProteinPilotTM Software (ABSciex, USA) using the ParagonTM and Pro GroupTM Algorithms and Uniprot 2015 database for Mus musculus. The false discovery rate (FDR) analysis was set to a confidence level of 95%. The label free statistical comparative analysis was performed using PeakViewTM Software (ABSciex, USA) with MS/MS(ALL) with SWATH™ Acquisition MicroApp 2.0 and MarkerViewTM (ABSciex, USA). Retention time alignment was obtained using selected peptides (top confidence and top level transitions) from top score protein of each analysis. Processing settings were: 7 peptides per protein, 7 transitions per peptide, 80% and 88% peptide confidence (according to Paragon algorithm result) respectively for secretome and microvesicles/exosomes and 5% FDR; XIC options: extraction window 10 min, width 50 ppm and 0.1 Da. Normalization was done using a global normalization of profiles (total protein content). Principal Component Analysis (PCA) was performed in order to evidence groupings among the data set. Groups were compared with t-test using fold change (FC) > 2 and p-value < 0.05.
Western blot analysis
Protein extracts (20 µg) were run on a 10% SDS-PAGE and transferred onto a nitrocellulose membrane (Amersham) using Trans-Blot SD Semi-Dry transfer cell (Biorad). The following antibodies were used: anti-osteopontin (anti-OPN, 1:500) (Meridian LifeScience); anti-CD44 (1:300) (Abcam); anti-chitinase 3-like 1 (anti-CHI3L1; 1:300) (Biorbyt).
BLM in vivo administration
Wild-type C57BL/6 female and male mice were exposed to BLM (Sigma-Aldrich, Milan, Italy) by intratracheal instillation. Mice were anesthetized with ketamine/xylazine (Virbac, srl) (ketamine 80 mg/kg intraperitoneal, xylazine 10 mg/kg intraperitoneal) and underwent intratracheal instillation of a single sub-lethal dose of 3 U/kg of BLM dissolved in 0.1 ml saline (0.9% NaCl). A control group underwent surgery for an intratracheal instillation of physiological saline solution. Mice were monitored for 28 days and then sacrificed.
Histological and immunohistochemical analyses
Mouse lungs were fixed by immersion in 5% buffered formalin for 7-10 days. Following standard histology processing, 10-20 sections, 5 μm thick, were cut from paraffin embedded samples by rotary microtome (Microm HM 300, Bio-optica) and stained with Masson trichrome (Bio-optica) for collagen.
For immunohistochemistry, sections were placed on positively charged slides, deparaffinized, rehydrated and washed in distilled water. After incubation in H2O2 at room temperature, antigen retrieval was accomplished (citrate buffer pH= 6 in microwave for 10 min at 500 W). The following primary antibodies were used: anti- α-smooth muscle actin (anti- α -SMA, 1:100) (Santa Cruz); anti-S100 calcium-binding protein A4 (anti-S100A4, 1:500) (Novus biologics); anti- vimentin (anti- VIM, 1:50) (Santa Cruz); anti-osteopontin (anti-OPN, 1:400) (Meridian Life Science); anti-chitinase 3-like 1 (anti-CHI3L1; 1:300) (Biorbyt). Omission of the primary antibody served as a negative control. Antibody binding to cells and/or to extracellular components is visible as brown or dark brown stain (DAB), negative cells are blue stained (haematoxylin counterstain). All sections were examined under light microscopy (Olympus BX43) and digitized by a video system (Olympus D20 camera) interfaced to Olympus Cell Sens Dimension software for image acquisition.
Results
Fibroblasts were isolated from lungs of healthy mice, cultured and treated with BLM, in order to induce and study the phenotypic changes that are likely connected with the fibrotic process.
To identify a dose of BLM able to determine phenotypic changes without inducing cell death, MTT assay was used and the drug was added to the cell culture in a specific range of concentration (from 0 µg/ml to 3.5 µg/ml) for up to 72 hours. Cell viability decreased very slowly and never reached values below 70%. It started to decrease with 1.5 µg/ml BLM and no differences were observed after 48 hours of treatment for all doses used. For these reasons, 1.5 µg/ml was chosen for successive experiments.
In order to demonstrate the switch of lung fibroblasts to an activated phenotype, also referred to as myofibroblasts, when treated with BLM, immunohistological analyses were performed and the expressions of αSMA, S100A4 and vimentin (VIM) were examined. The expression of α-SMA (notoriously neo-expressed by myofibroblasts), and S100A4, which is a marker of pulmonary fibrosis increased in BLM-treated cells, while VIM (recognized marker for fibroblasts) was more expressed in untreated cells (Figure 1). These observations strengthened the hypothesis that primary cultures of lung fibroblasts treated with BLM can represent an in vitro model of lung fibrosis.
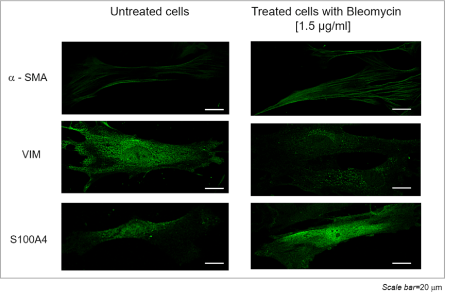
Figure 1. Lung fibroblast immunocytochemistry: α-SMA and S110A4 in BLM untreated (left) and treated (right) fibroblasts (Scale bar=20µm)
For secretome analysis, fibroblasts were cultured until cell confluence reached 90% and incubated in serum-free medium supplemented or not with BLM for 48 hours. Mass spectrometry analysis allowed the identification of 263 proteins (Table S1). In Figure 2, the subcellular localization of these proteins is reported according to UniProt consortium (http://www.uniprot.org). In this specific setting, it is worth of note that 44% of proteins are exosome, matrix and extracellular space proteins.
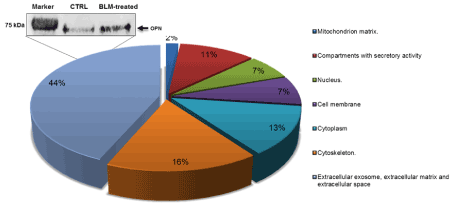
Figure 2. Subcellular location of identified secretome proteins. Protein classification was made according to UniProt consortium http://www.uniprot.org. Western blot was carried out to validate secretome analyses for OPN expression in BLM-treated and control (CTRL) fibroblasts
Among the 263 identified proteins, 17 resulted differentially expressed in BLM-treated fibroblasts compared to untreated cells. These proteins, their p-values and fold changes are reported in (Table S2). Table 1 shows the differentially expressed proteins that, according to literature, may be associated to the fibrotic process.
Table 1. Differentially secreted proteins
Groups |
DOWN regulated proteins
BLM vs CONTROL |
UP regulated proteins
BLM vs CONTROL |
Chaperones |
Heat shock protein HSP 90-alpha |
|
Actin-binding proteins and cytoskeleton |
Cadherin-2 |
|
Extracellular matrix, MMPs and related factors |
Lysyl oxidase homolog 2 |
MMP-2
14-3-3 protein
MMP-3
Osteopontin |
Inflammation and oxidative stress |
|
Legumain
Chitinase-3-like protein 1 |
TGF-β |
Calreticulin |
|
Proteins are grouped according to their putative involvement in lung fibrosis (p-value<0.05; Fold Change (FC) >2 for up-regulated and 1/FC >2 for down-regulated proteins)
To validate proteomics results, western blots were performed using anti-OPN antibody. OPN was found to be up-regulated in secretome of BLM-treated lung-fibroblasts as compared to the secretome of untreated cells (CTRL), confirming mass spectrometry based data.
The major drawback of secretome assessment is represented by the necessity to culture the cells in serum-free medium, a state of starvation and thus stress, due to the presence in serum of albumin and other abundant proteins that mask less abundant secreted proteins. In order to assess secreted proteins in more optimal physiological conditions, we isolated and studied extracellular vesicles (EV), i.e microvesicles and exosomes from BLM-treated lung fibroblasts cultured in standard conditions, in the presence of serum.
This protocol allowed the identification of up to 464 proteins (Table S3). Identified proteins were grouped according to their subcellular localization (Figure 3).
Forty-four proteins resulted differentially expressed and were analyzed according to their functions and activity, playing particular attention to their potential involvement in pulmonary fibrosis. The complete list is stated in (Table S4). In Table 2 were reported the differentially expressed proteins that can be associated to fibrosis according to the literature.
Table 2. Differentially expressed proteins in released microvesicles
Groups |
DOWN regulated proteins
BLM vs CONTROL |
UP regulated proteins
BLM vs CONTROL |
Actin-binding proteins and cytoskeleton |
Gelsolin |
Septin-2
Filamin-C
Keratin, type II cytoskeletal 1 |
Extracellular matrix and related factors |
|
Procollagen C-endopeptidase
enhancer 1
Protein-tyrosine kinase 2-beta |
Oxidative stress and redox regulation |
Flavin reductase
Peroxiredoxin-2
Peroxiredoxin-6 |
|
TGF-β |
Chloride intracellular channel protein 4
Pigment epithelium-derived factor
Galectin-1
Coagulation factor V
CD44
Transgelin-2 |
|
DNA repair |
Nucleophosmin |
|
Inflammation |
Carboxypeptidase N catalytic chain
Cdc42
Carboxypeptidase B2 |
|
Calcium-binding proteins |
Calmodulin
Calponin |
|
Proteins are grouped according to their putative involvement in lung fibrosis (p-value<0.05; Fold Change (FC) >2 for up-regulated and 1/FC >2 for down-regulated proteins)
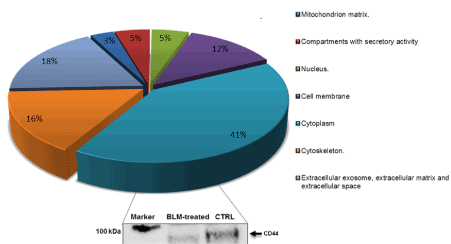
Figure 3. Subcellular localization of total identified vesicle enclosed proteins. Protein classification was made according to UniProt consortium, http://www.uniprot.org. Western blot was carried out to validate proteomics data for CD44 expression in BLM-treated and control (CTRL) fibroblasts
Western blot with anti-CD44 antibody was performed to validate the proteomics results; data shown in Figure 3 confirm a down-regulation of this protein in BLM-treated cells.
In order to investigate the possible transferability of the in vitro results to the in vivo animal model, mice were exposed to BLM (3 U/kg) by intratracheal instillation, while a control group was treated with PBS. Animals were sacrificed after 28 days and lungs processed for histological analyses. In BLM-treated lungs, inflammatory infiltrates were evidenced and a marked production and accumulation of collagen fibers were detected in Mallory’s trichrome stain. Onset and progression of fibrosis was confirmed by immunohistochemical images showing an increase of positivity to both anti-α-SMA and anti-S100A4 in BLM-treated tissue compared with untreated one (Figure 4).
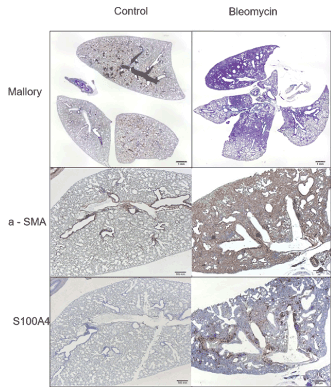
Figure 4. Histological and immunohistochemical tissue characterization of BLM-induced lung fibrosis. Lung sections from mice treated with 3 U/Kg of bleomycin were compared with sections from physiological saline solution-treated mice. Representative micrographs of lung tissue stained with Mallory’s trichrome (Mallory) show focal areas of interstitial fibrosis in treated lungs (Scale bar=1 mm). Representative images for α-SMA and S100A4 show positive areas of fibrosis in BLM-treated lungs. (Scale bar=500 µm)
Immunohistochemistry analyses were also performed with antibodies specific for two differentially secreted proteins, osteopontin (anti-OPN) and chitinase-3-like protein 1 (anti-CHI3L1) (Figure 5) both proteins were strongly expressed in treated fibrotic lungs, confirming and validating the proteomics results.
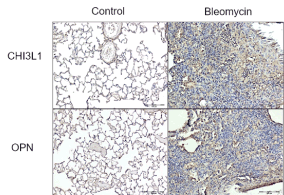
Figure 5. Representative photomicrographs of immunostained lung sections. Lung sections from mice treated with 3 U/Kg of bleomycin were compared with sections from physiological saline solution-treated mice. Representative images for OPN and CHI3L1 antibodies show positive areas of fibrosis in BLM-treated lungs. (Scale bar=500 µm)
Discussion
Hallmarks of fibrosis progression and worsening are represented by hyper-proliferation of type II alveolar epithelial cells and recruitment and proliferation of fibroblasts, leading to an increase of fibroblastic foci. Activated fibroblasts are largely responsible for the production and deposition of interstitial collagen and other ECM components [10,11]. Specifically, when tissues are injured, locally residing connective fibroblasts acquire contractile activity by forming contractile stress fibers, neo-expressing α-SMA, while VIM expression decreases. In fact α-SMA expressing myofibroblasts not only promote contraction but also synthetize high levels of both extracellular matrix components and matrix degrading proteases playing a fundamental role in remodeling injured tissue [12-17]. A novel marker of pulmonary fibrosis is the fibroblast-specific protein 1 (FSP1) also identified as S100A4. Normal murine lung parenchyma expresses very low levels of S100A4, but its levels rapidly increase during BLM-induced pulmonary fibrosis. S100A4 expression is reported to correlate with collagen deposition and lung parenchyma architectural changes in the murine model of lung fibrosis. As evidenced in Figure 1 lung fibroblasts treated with BLM acquired the morphological and immunohistochemical features that characterize fibrosis. The main focus of this study is represented by the modulation of protein synthesis and release in the BLM-induced phenotype switch from fibroblasts to myofibroblasts. As a matter of fact, the activation and proliferation of lung fibroblasts precede accumulation of ECM components in the damaged alveolus and it is a key step in the transformation from a potentially reversible disorder to a progressive and irreversible one. Indeed, the amount of fibroblast/myofibroblasts foci is considered a main prognostic factor in pulmonary fibrosis disease [11].
In order to gain better insight in the fibrotic process, we analyzed the proteins released by BLM-activated fibroblasts. Secreted proteins play important roles in the regulation of many physiological processes via paracrine/autocrine mechanisms and they gained interest as potential biomarkers and therapeutic targets of diseases [18,19]. Moreover, the identification of proteins released in the extracellular space by fibrotic cells may be of great interest for the comprehension of lung fibrosis, being characterized by increased collagen deposition, accumulation of extracellular matrix components, cell proliferation/trans-differentiation and alteration of normal lung structure. 263 secreted proteins were identified, 44% are exosome, matrix and extracellular space proteins and among them 17 resulted differentially expressed in BLM-treated fibroblasts compared to untreated cells. Several up-regulated proteins are factors related to ECM; it is worth of note that ECM plays a critical role in the repair process, orchestrating the transitions of cell phenotype and function [7,20].
Among the up-regulated proteins, osteopontin (OPN) is particularly interesting since recent studies reported that after BLM instillation, OPN -/- mice showed reduced lung fibrosis with decreased active transforming growth factor-beta 1 (TGF-β1) and reduced type I and type III collagen expression [21-23]. OPN also influences ECM remodeling by the alteration of collagen matrix and the induction of several matrix metalloproteinases (MMPs), such as MMP-1, MMP-2 and MMP-9 [24]. It has long been accepted that MMPs play an important role in the pathogenesis of pulmonary fibrosis; as a matter of fact, we identified two up-regulated MMPs in BLM-treated fibroblasts: MMP-2 and MMP-3. MMP-1, MMP-2, MMP-7, and MMP-9 have been found highly expressed in pulmonary fibrosis and, in particular, in fibroblasts foci [25]. MMPs, including MMP-3, have the ability to regulate tissue repair altering the activity of non-matrix proteins, such as cytokines and membrane receptors, leading to the activation of pathways responsible of epithelial-mesenchymal transformation (EMT) [26]. As a consequence, MMP-3 could play a role in the origin of myofibroblasts [11]. Many factors and proteins interact with MMPs modulating their function and, among the others, the 14-3-3 proteins, a class of highly conserved molecular chaperones, represent important MMP-1 regulators. To close this interesting circle, it is of note that we also reported up-regulation of chitinase 3-like 1 (CHI3L1), that is involved in the inhibition of degradation of type I collagen by MMP-1 [27-29].
Secreted vesicles represent a growing field of interest from a diagnostic point of view, aiming at identifying disease biomarkers in biological fluids (such as blood, urine, saliva) [9,30-32]. In the present study, the isolation of released vesicles allowed the identification of up to 464 proteins.
Forty-four proteins resulted differentially expressed and among them both peroxiredoxin 6 (Prdx6) and peroxiredoxin 2 (Prdx2) resulted down-regulated in BLM-treated fibroblasts suggesting a role for the redox system in fibrotic process. Peroxiredoxins are antioxidant enzymes and interestingly, Prdx2 was down-regulated in fibroblasts foci of idiopathic pulmonary fibrosis when compared to healthy control lungs [33].
Prdx6 is expressed at high levels in the lung where it has been recognized as a novel antioxidant enzyme that may play a critical role against oxidative damage [34]. Wang and colleagues demonstrated that transgenic mice overexpressing Prdx6 showed an increased resistance to lung injury in hyperoxia, leading to a major degradation of hydrogen peroxide (H2O2) and conferring protection against several forms of oxidative stress. As a consequence, the decreased enzyme expression should potentiate lung injury [35-38].
Interestingly, several differentially expressed proteins are related to the TGF-β pathway since TGF-β is a potent fibrotic factor, an important activator of phenotype trans-differentiation from fibroblasts to myofibroblasts and its activity can be associated with the deposition and remodeling of ECM by stimulating cells to increase matrix protein synthesis, decrease the production of matrix-degrading proteases and modulate integrin expression [39]. CD44 is one of the proteins potentially involved in TGF-β pathway modulation, interacting with cytoskeletal components and other ligands, such as OPN, collagen and MMPs, influencing the adhesion and motility of fibroblasts, thereby supporting their potential role in tissue remodeling and fibrosis [40-42]. Several studies suggested that the expression of CD44 play a major role in the resolution of inflammation and transition to reparative fibrosis in response to tissue injury.
The phenotypic change towards an activated state is necessarily related to remodeling of actin cytoskeleton; in fact, several actin-binding proteins were found differentially regulated in our samples. Among them, we found gelsolin (GSN) and transgelin-2 (Tagln-2) down-regulated in released vesicles from treated fibroblasts. This result is in line with the already published data in various types of cancer, rheumatoid arthritis and pulmonary fibrosis [43-45].
The different forms of pulmonary fibrosis in humans, in particular IPF, are burdened by high morbidity and mortality. Although pathogenic pathways and mediators of lung fibrosis have been in depth examined in the last few years, the molecular pathways involved are not fully understood so far. In animals treated with BLM, lung histology shows features similar to those present in humans with IPF. To provide a deeper insight in lung fibrotic pathogenetic mechanisms, proteomic analyses were undertaken on BLM-treated lung fibroblasts. Lung fibrosis is characterized by ECM synthesis and remodeling, therefore we focused on the study of secretome and microvesicle enclosed proteins. Several MMPs and ECM proteins resulted up-regulated in secretome of treated fibroblasts, as well as OPN, which plays a role in both inflammation and tissue repair. The relevance of OPN in phenotype modulation of fibroblasts is confirmed by vesicle analyses that evidenced the differential expression of CD44, which interacts with OPN and MMPs and influences the inflammatory response. An increased expression of OPN and CHI3L1 in the fibrotic lung tissue in an animal model of BLM-induced fibrosis validated our proteomics results suggesting the possibility to translate in vitro data to in vivo settings.
In conclusion, in an experimental model of BLM-induced lung fibrosis, several key proteins have been identified. The paucity of current clinical treatment represents a strong incentive to further research in order to assess whether these factors may be considered as potential targets for novel therapeutic approaches.
Authors contributions
Conception and design, VDL, AC, MAM. Laboratory testing and data collection, VDL, AC, SR, LC, NU, CDP, MT, GP, SB. Data analysis and interpretation: VDL, AC, MAM, SR, GP. Drafting and revising of manuscript: VDL, AC, MAM. All authors read and approved the final manuscript.
Funding information
This work was supported by a research grant of the Institute of Clinical Physiology (IFC), National Research Centre (CNR), Pisa, Italy.
Competing interests
The authors declare that they have no competing interests.
View supplementary data
References
- Pardo A, Selman M (2002) Molecular mechanisms of pulmonary fibrosis. Front Biosci 7: 1743-1761. [Crossref]
- Ahmad K, Nathan SD (2018) Novel management strategies for idiopathic pulmonary fibrosis. Expert Rev Respir Med 30: 1-12. [Crossref]
- Moeller A, Rodriguez-Lecompte JC, Wang L, Gauldie J, Kolb M (2006) Models of pulmonary fibrosis. drug discovery today. Disease Models 3: 243-249.
- Walters DM, Kleeberger SR (2008) Mouse models of bleomycin-induced pulmonary fibrosis. Curr Prot in Pharmacol 5.46: 1-17. [Crossref]
- Della Latta V, Cecchettini A, Del Ry S, Morales A (2015) Bleomycin in the setting of lung fibrosis induction: from biological mechanisms to counteractions. Pharmacological Research 97: 122-130. [Crossref]
- Selman M, King TE, Pardo A (2001) Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 134: 136-51. [Crossref]
- Selman M, Pardo A (2002) Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res 3: 3. [Crossref]
- Thannickal VJ, Toews GB, White ES, Lynch JP 3rd, Martinez FJ (2004) Mechanisms of pulmonary fibrosis. Annu Rev Med 55: 395-417. [Crossref]
- Comelli L, Rocchiccioli S, Smirni S, Salvetti A, Signore G, et al. (2014) Characterization of secreted vesicles from vascular smooth muscle cells. Mol Biosyst 10: 1146-1152. [Crossref]
- Lawson WE, Polosukhin VV, Zoia O, Stathopoulos GT, Han W, et al. (2005) Characterization of fibroblast-specific protein 1 in pulmonary fibrosis. Am J Respir Crit Care Med 171: 899-907. [Crossref]
- Rocha-Azevedo B, Ho CH, Grinnell F (2015) PDGF-stimulated dispersal of cell clusters and disruption of fibronectin matrix on three-dimensional collagen matrices requires matrix metalloproteinase-2. Mol Biol Cell 26: 1098-1105.
- Scotton CJ, Chambers RC (2007) Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest 132: 1311-1321. [Crossref]
- Gabbiani G (2004) The evolution of the myofibroblast concept: a key cell for wound healing and fibrotic diseases. G GERONTOL 52: 280-282.
- Gabbiani G (2003) The myofibroblast in wound healing and fibrocontractive diseases. J Pathol 200: 500-503. [Crossref]
- Wipff PJ, Hinz B (2009) Myofibroblasts work best under stress. J Bodyw Mov Ther 13: 121-127. [Crossref]
- Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C (2001) Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell 12: 2730-2741.
- Grinnell F (2003) Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol 13: 264-269.
- Strutz F, Okada H, Lo CW, Danoff T, Carone RL, et al. (1995) Identification and characterization of a fibroblast marker: FSP1. J Cell Biol 130: 393-405. [Crossref]
- Hathout Y (2007) Approaches to the study of the cell secretome. Expert Rev Proteomics 4: 239-248. [Crossref]
- Raimondo F, Morosi L, Chinello C, Magni F, Pitto M (2011) Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics 11: 709-720. [Crossref]
- Thannickal VJ, Henke CA, Horowitz JC, Noble PW, Roman J, et al. (2014) Matrix biology of idiopathic pulmonary fibrosis: a workshop report of the national heart, lung, and blood institute. Am J Pathol 184: 1643-51.
- Takahashi F, Takahashi K, Okazaki T, Maeda K, Ienaga H, et al. (2001) Role of osteopontin in the pathogenesis of bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 24: 264-271. [Crossref]
- Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, et al. (2005) Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2: e251. [Crossref]
- O'Regan A (2003) The role of osteopontin in lung disease. Cytokine Growth Factor Rev 14: 479-488. [Crossref]
- Pardo A, Selman M (2006) Matrix metalloproteases in aberrant fibrotic tissue remodeling. Proc Am Thorac Soc 3: 383-388. [Crossref]
- Asdaghi N, Kilani RT, Hosseini-Tabatabaei A, Odemuyiwa SO, Hackett TL, et al. (2012) Extracellular 14-3-3 from human lung epithelial cells enhances MMP-1 expression. Mol Cell Biochem 360: 261-270. [Crossref]
- Yamashita CM, Dolgonos L, Zemans RL, Young SK, Robertson J, et al. (2011) Matrix metalloproteinase 3 is a mediator of pulmonary fibrosis. Am J Pathol 179: 1733-1745. [Crossref]
- Ober C, Tan Z, Sun Y, Possick JD, Pan L, et al. (2008) Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 358: 1682-1691. [Crossref]
- Furuhashi K, Suda T, Nakamura Y, Inui N, Hashimoto D (2010) Increased expression of YKL-40, a chitinase-like protein, in serum and lung of patients with idiopathic pulmonary fibrosis. Respir Med 104: 1204-1210.
- Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, et al. (2011) Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 73: 479-501. [Crossref]
- Simpson RJ, Jensen SS, Lim JW (2008) Proteomic profiling of exosomes: current perspectives. Proteomics 8: 4083-4099. [Crossref]
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D'Souza-Schorey C (2010) Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 123: 1603-11
- Lakkaraju A, Rodriguez-Boulan E (2008) Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol 18: 199-209. [Crossref]
- Vuorinen K, Ohlmeier S, Leppäranta O, Salmenkivi K, Myllärniemi M, et al. (2008) Peroxiredoxin II expression and its association with oxidative stress and cell proliferation in human idiopathic pulmonary fibrosis. J Histochem Cytochem 56: 951-959.
- Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, et al. (2006) Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A 103: 13180-13185.
- Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB (2004) Lung injury and mortality with hyperoxia are increased in peroxiredoxin 6 gene-targeted mice. Free Radic Biol Med 37: 1736-1743. [Crossref]
- Wang Y, Phelan SA, Manevich Y, Feinstein SI, Fisher AB (2006) Transgenic mice overexpressing peroxiredoxin 6 show increased resistance to lung injury in hyperoxia. Am J Respir Cell Mol Biol 34: 481-486.
- Kinnula VL, Fattman CL, Tan RJ, Oury TD (2005) Oxidative stress in pulmonary fibrosis: a possible role for redox modulatory therapy. Am J Respir Crit Care Med 172: 417-422. [Crossref]
- Day BJ (2008) Antioxidants as potential therapeutics for lung fibrosis. Antioxid Redox Signal 10: 355-370. [Crossref]
- Honda E, Park AM, Yoshida K, Tabuchi M, Munakata H (2013) Myofibroblasts: Biochemical and proteomic approaches to fibrosis. Tohoku J Exp Med 230: 67-73. [Crossref]
- Acharya PS, Majumdar S, Jacob M, Hayden J, Mrass P, et al. (2008) Fibroblast migration is mediated by CD44-dependent TGF beta activation. J Cell Sci 121: 1393-1402. [Crossref]
- Evanko SP, Potter-Perigo S, Petty LJ, Workman GA, Wight TN (2015) Hyaluronan controls the deposition of fibronectin and collagen and modulates TGF-β1 induction of lung myofibroblasts. Matrix Biol 42: 74-92. [Crossref]
- Li Y, Jiang D, Liang J, Meltzer EB, Gray A, et al. (2011) Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med 208: 1459-1471.
- Maniatis NA, Harokopos V, Thanassopoulou A, Oikonomou N, Mersinias V, et al. (2009) A critical role for gelsolin in ventilator-induced lung injury. Am J Respir Cell Mol Biol 41: 426-432. [Crossref]
- Oikonomou N, Thanasopoulou A, Tzouvelekis A, Harokopos V, Paparountas T, et al. (2009) Gelsolin expression is necessary for the development of modelled pulmonary inflammation and fibrosis. Thorax 64: 467-475. [Crossref]





