Atherosclerosis is a cardiovascular disease marked by chronic inflammation. The disease is intimately related to the accumulation of fatty acids in the tunica intima of medium and large caliber arteries. The development of the disease respond to alteration of the vascular and immune system homeostasis. The inflammatory process is aggravated by the action of immune cells. The diagnosis of the disease is invasive in most cases. Thus, new diagnostic and therapeutic techniques are required and could increase the quality of life of atherosclerotic patients. The eNOS gene has been indicated as an efficient biomarker of cardiovascular diseases, including atherosclerosis, stroke and myocardium infarction. The eNOS gene and the protein it encodes control the production of nitric oxide. This compound takes part in a large variety of biological functions and its action is crucial for the protection of arteries against damage. Here, an in-silico approach was used to design two small active peptides as a way to stabilize eNOS and guarantee its normal level of function regarding the production of nitric oxide. We hypothesize that the use of designed peptide would lead to an efficient therapy and with less or none side effect to atherosclerotic patients, as a substitution of statins therapy, such as atorvastatin. For future perspective, an assay of the modulating peptides is being designed to be tested in vitro and in vivo in order to be possibly used as a new therapeutic agent.
eNOS, atherosclerosis, bioinformatics, modulating peptides
Atherosclerosis is a common worldwide disease marked by a chronic inflammatory process due to the accumulation of fatty acids in the tunica intima. The disease leads to the formation of an atheromatous plaque through the accumulation and deposition of platelet factors, fatty acids, immune system and smooth muscle cells. The direct consequence of such factor is the narrowing of the arterial lumen. Eventually, the atheromatous plaque undergoes fibrosis and calcification, which over time might lead to severe symptoms and even death [1].
The onset of the atherosclerosis depends on alteration of the vascular and immune system homeostasis [2]. Cells from the immune system affect the pathogenesis of the disease aggravating inflammatory processes within the endothelium [3]. Atherosclerosis is a silent disease and takes a long period to manifest due to genetic, physiological and environmental factors [4]. In addition, cardiovascular diseases, such as atherosclerosis, are the leading cause of death worldwide [5]. Several risk factors are related the development and manifestation of such diseases.
Diagnosis of atherosclerosis is usually invasive. The necessity of finding new and efficient methods of identifying individuals predisposed to develop atherosclerosis is real [1]. Bioinformatic assays have been trying to accomplish that task along with more efficient and less invasive ways to treat cardiovascular diseases [6]. The incredibly complex genetic trait presented by atherosclerosis makes the development of such methods difficult and time-consuming [2].
One of the most important genes investigated as a biomarker of cardiovascular diseases is eNOS (endothelial nitric oxide synthase). The eNOS coding gene is located on chromosome 7 and is responsible for the production of nitric oxide. This compound has lipophilic properties and participates in many intracellular homeostatic processes [7–9]. Nitric oxide also controls the vascular tone [10], cell proliferation [11], adhesion of immune system cells to arteries wall undergoing inflammation processes [12] and platelet aggregation [13]. Mutation within the eNOS gene coding sequence and within the amino acid sequence of the eNOS protein may affect endothelial function [14]. The unavailability of nitric oxide in proper levels increase the production and release of reactive oxygen species (ROS) that cause endothelial injury and is related to several cardiovascular diseases, such as atherosclerosis [15], coronary artery disease [16], diabetes [17] and infarction [18].
Statin drugs are widely used in order to prevent cardiovascular diseases in patients at high risk of developing such diseases [19]. Atorvastatin is a common type o statin used to treat patients with high levels of lipids. The most common side effects are nausea, diarrhea, heart burn, joint and muscle pains, rhabdomyolysis, liver and kidney problems and diabetes [20]. Similar to general statins, atorvastatin inhibits HMG-CoA reductase (3-hydroxy-3-methyl-glutaryl-coenzyme A reductase) that takes part in cholesterol production.
Here, we used an in-silico approach to predict modulator peptides of the eNOS protein in order to enhance nitric oxide production. Similar effect is achieved by atorvastatin therapy in patients suffering from atherosclerosis. We hypothesize that the use of designed peptide would lead to an efficient therapy and with less or none side effect to atherosclerotic patients. For future perspective, an assay of the modulating peptides is being designed to be tested in vitro and in vivo in order to be possibly used as a new therapeutic agent.
The prediction of the three-dimensional structure of eNOS was modeled by the I-TASSER (Iterative Threading Assembly Refinement) server [21]. The modeling process was based on homologous templates of protein structures resolved previously and deposited in the PDB (protein databank). The predicted eNOS structure underwent fold recognition via Monte Carlo computational simulations and the secondary structure of the protein was determined by PSSpred (Protein Secondary Structure Prediction) [22,23]. The best ranked predictions was clustered by means of conformation and energy using SPICKER [24] in order to determine the nearest native structures and molecular dynamics was used as a refinement.
The visualization software PyMol (https://pymol.org) was used in order to analyze the eNOS structure, to highlight polymorphic residues and finally to design peptides to modulate the eNOS conformation and function. Amino acid residues that significantly contribute to the free-energy of binding and stability of the interaction between protein and peptide were predicted by the KFC2 [25] server. The identification of such amino acid residues depends on physical and chemical properties of the environment around those amino acids. Alanine scanning mutagenesis are considered for the prediction of hot spots in the protein structure. The hot spot scores depend on conformation (score a) and on chemical properties (score b). Clinically important polymorphic residues for the eNOS and p53 proteins were identified through the dbSNP (database of single nucleotide polymorphism).
Both the gene and the protein eNOS control the nitric oxide production. The release of nitric oxide affects positively several physiological conditions [26], such as vascular tone [27], apoptosis [28] and platelet aggregation [29] and diseases, such as several types of cancer [30] and vascular diseases [31,32]. Researchers have been using in silico approaches in order to contribute to the development of novel diagnostic and therapeutic methods [33,34]. The prediction of important amino acid residues acting as hot spots within the binding pocket of protein interacting with drugs has shed some light on the design of modulators peptides of the function and conformation of target proteins [34-36]. Polymorphisms in regions of hot spot clusters alter the stable conformational of proteins and may raise the predisposition to cardiovascular diseases [37]. The present bioinformatics approach allowed the design of two modulator peptides (peptide 185 and peptide 207) that stabilize the eNOS protein and its function in atherosclerosis.
Figure 1 shows the predicted hot spots on eNOS structure (Table 1) regarding the designed peptides. The prediction was based on three types of interaction energy (electrostatic, hydrophobic and Van der Waals) that regulate stabilization of the protein-peptide complex. Electrostatic forces have been shown to stabilize protein and protein complexes conformation [38,39] and might affect diseases onset and progression [40]. In addition, interactions mostly based on hydrophobicity and Van der Waals are related to stabilization of protein and ligands in a way that they could enhance response to statins and improve prognosis of the cardiovascular diseases [41].
Table 1. Predicted hot spot amino acid residues that contribute to the stabilization of the eNOS protein in order to maintain its function and nitric oxide production
Hot Spot Residue* |
Structure Score |
Chemical Score |
R170 |
0.36 |
0.04 |
F105 |
0.63 |
0.04 |
W244 |
1.38 |
0.29 |
W322 |
1.46 |
0.25 |
L326 |
0.6 |
0.14 |
Q476 |
0.52 |
0.06 |
D478 |
1.37 |
0.01 |
W480 |
1.28 |
0.29 |
R474 |
0.99 |
0.14 |
*Prediction was performed via KFC2 [25].
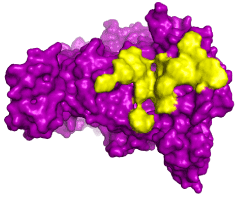
Figure 1. The eNOS protein and the predicted hot spots according to the binding of modulating peptides. The prediction of hot spot residues (yellos) on the eNOS structure (purple) was based upon interaction with modulating peptides designed to stabilize the conformation of the protein according to its ability to regulate production and release of nitric oxide. The same effect is achieved by the use of statins such as atorvastatin
Usually, hot spots occurs in clusters and the design of modulating peptides is performed around these regions in order to allow interaction between protein-drug in a more stable way [42]. The hot spots, in the present work, were found to form clusters, as it usually is, and contribute to the stability of the interaction between eNOS and peptide (Tables 1).
The residue R70 and R474 were predicted as a hot spot (Table 1). Arginine (R) is an amphipathic residue and is frequently present on the surface of proteins, allowing its interaction with other organic molecules. This fact makes this amino acid play important roles as hot spots in several proteins related to diseases [43]. The R residues are involved in polar bonding between proteins and target molecules, such as drugs and peptides. In addition, this amino acid is often interacting with residues of several other proteins through electrostatic forces [44], working as an electrostatic adhesive force when organic molecules interact with each other [45]. In our assay, the R hot spot residues establish polar interactions with residues in the vicinity within the same polypeptide chain and with amino acids from the designed peptide (peptide 185) (Figure 2A). Therefore, R70 as a hot spot residue is essential for the stability of eNOS, establishing contacts with peptide 185 through bonds in the range of 4 Å (Figure 2B).
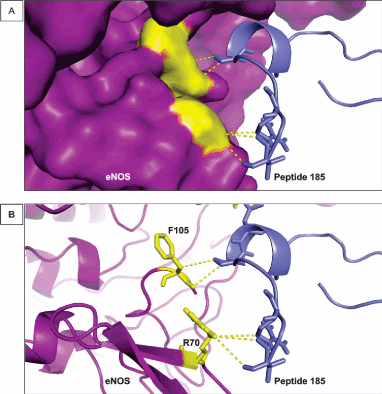
Figure 2. Interaction between R70 and F105 of eNOS and peptide 185. (A) The image shows the surface of eNOS in purple and the hot spot predicted residues by KFC2 [25] in yellow. Peptide 185 is represented in blue. This small molecule helps to stabilize eNOS conformation and maintain its activity. (B) The cartoon representation of eNOS showing the structures of R70 and F105 and their interaction with peptide 185. The distance of the bonds established by R70 and F105 in in the range of 4 and 3 Å, respectively
The residues Q (glutamine) 476 (Figure 3), D (aspartate) 478 and W (tryptophan) 480 also help to stabilize the conformation of eNOS interaction with peptides. Figure 3A shows the disposition of the peptide near this cluster of residues. Histidine residues also form this cluster of hot spots (data not shown) and significantly reduce the free-energy of interaction, playing an important role on the biological function that proteins are involved in [46,47]. Histidine residues exert a variety of roles in molecular interactions performed by protein in general due to their biochemical properties. Histidine are able to modulate the electrostatic coefficient of positively or negatively charged amino acids. This ability is crucial for the stability of proteins [48] and for designing therapeutic peptides [49]. In addition, leucine and tryptophan residues (defined as hot spots in the binding of eNOS and small molecules or drugs such as statins) contribute to the stability of protein conformation and binding energy through polar interaction and this property has also been demonstrated to occur for histidine residues [48,49].
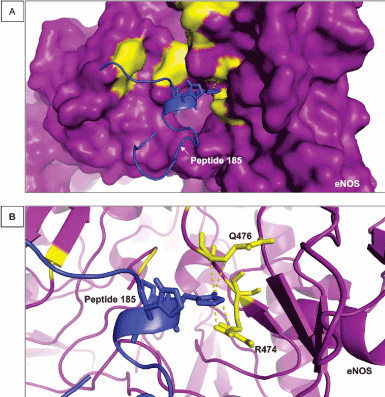
Figure 3. Interaction between R474 and Q476 of eNOS and peptide 185. (A) The image shows another view of the surface of eNOS in purple and the hot spot predicted residues in yellow. Peptide 185 is represented in blue. (B) The cartoon representation of eNOS showing the structures of R474 and Q476 when they are interacting directly with peptide 185. The distance of the bonds established by R474 and Q476 is in the range of 3 Å
R474 and Q476 residues contribute to the free-energy of stabilization of eNOS and binding to the peptide 183 mainly through electrostatic-favored bonds. Hot spot residues that perform those types of interactions play important roles on the intermolecular association and aggregation of a protein and a binding molecule via polar bonds [50] in a range of approximately 3 Å. In addition, the amino acid aspartate participates in interactions among several types of organic molecules, such as peptides, proteins and RNAs [51]. Tryptophan, another hot spot identified in the present approach (Table 1), has been pointed as a common component of protein-ligand interfaces, anchoring the structure of the binding organic molecules [52] and stabilizing binding sites of proteins [53].
A tryptophan residue classified as a hot spot residues by our analysis contribute to the free energy of eNOS binding to a second designed peptide (peptide 207) through hydrophobic interactions. Hydrophobic residues, such as tryptophan, and their bonding to neighboring residues help to maintain eNOS folding, mainly because of their hydrophobic properties and tendency to be placed within cavities of the protein structure, away from the solvent [54]. Figure 4A shows the disposition of peptide 207 and its placement in a protein cleft and Figure 4B shows how the hot spot D478 and W480 residues interact with neighboring residues of peptide 207. Tryptophan and establishes several polar interaction with peptide 207 in the range of 3 Å.
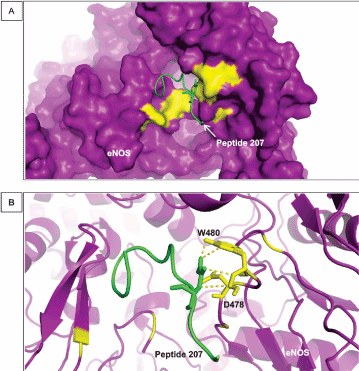
Figure 4. Interaction between D478 and W480 of eNOS and peptide 207. (A) The image shows a general view of the surface of eNOS in purple and the hot spot predicted residues in yellow linked to peptide 207 (green). (B) The cartoon representation of eNOS showing the structures of D478 and W480 when they interact directly with peptide 207. The distance of the bonds established those residues is in the range of 3 Å
Moreover, hydrophobic amino acid residues influence refolding and stability of secondary beta‐sheets [55-58]. Here, W322 is in a beta‐sheet chain and we hypothesize that it significantly contributes to the stabilization of the conformation of eNOS, mimicking the effects of statins, used as treatment of atherosclerosis. Single nucleotide polymorphisms that substitute this amino acid residue shows clinical relevance as it increase susceptibility to cardiovascular disease reducing the level of nitric oxide available for the intracellular and vascular homeostasis.
To our knowledge, no study aimed at the design of small molecules for the substitution of well-established atherosclerosis statin therapy, although several studies have tried to design modulator peptides of the eNOS protein [60-65].
Cardiovascular diseases are a leading cause of death globally. Eating and drinking habits, genetic and environmental factors are closely related to the susceptibility to infarction, atherosclerosis and hypertension. Bioinformatic approaches has invested in the modeling of protein structures, networks of interaction and design of small active molecules in order to improve diagnostic and therapy techniques. Here, an in-silico approach was used to design two small active peptides as a way to stabilize eNOS and guarantee its normal level of function regarding the production of nitric oxide. We hypothesize that the use of designed peptide would lead to an efficient therapy and with less or none side effect to atherosclerotic patients, as a substitution of statins therapy, such as atorvastatin. For future perspective, an assay of the modulating peptides is being designed to be tested in vitro and in vivo in order to be possibly used as a new therapeutic agent.
- Libby P, Bornfeldt KE, Tall AR (2016) Atherosclerosis: Successes, Surprises, and Future Challenges. Circ Res 118: 531-534.
- Rahman MS, Woollard K Atherosclerosis Adv Exp Med Biol 1003, 121-144 (2017)
- Gimbrone MA, García-Cardeña G (2016) Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res 118: 620-636.
- Head T, Daunert S, Goldschmidt-Clermont PJ (2017) The Aging Risk and Atherosclerosis: A Fresh Look at Arterial Homeostasis. Front Genet 8.
- https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(16)31678-6/fulltext
- Wang H, Liu D, Zhang H (2019) Investigation of the Underlying Genes and Mechanism of Macrophage-Enriched Ruptured Atherosclerotic Plaques Using Bioinformatics Method. J Atheroscler Thromb 26: 636-658.
- Dellamea BS, Leitão CB, Friedman R, Canani LH (2014) Nitric oxide system and diabetic nephropathy. Diabetol Metab Syndr 6: 17.
- Nakanishi K, Koshiyama T, Iba S, Ohba M (2015) Lipophilic ruthenium salen complexes: incorporation into liposome bilayers and photoinduced release of nitric oxide. Dalton Trans 44: 14200-14203.
- Fan Y, Lu H, Liang W, Hu W, Zhang J, et al. (2017) Krüppel-like factors and vascular wall homeostasis. J Mol Cell Biol 9: 352-363.
- Zhao Y, Vanhoutte PM, Leung SWS (2015) Vascular nitric oxide: Beyond eNOS. J Pharmacol Sci 129: 83-94.
- Tousoulis D, Kampoli AM, Tentolouris C, Papageorgiou N, Stefanadis C (2012) The role of nitric oxide on endothelial function. Curr Vasc Pharmacol 10: 4-18.
- Gutsaeva DR, Montero-Huerta P, Parkerson JB, Yerigenahally SD, Ikuta T, et al. (2014) Molecular mechanisms underlying synergistic adhesion of sickle red blood cells by hypoxia and low nitric oxide bioavailability. Blood 123: 1917-1926.
- Radziwon-Balicka A, Lesyk G, Back V (2017) Differential eNOS-signalling by platelet subpopulations regulates adhesion and aggregation. Cardiovasc Res 113: 1719-1731.
- Godo S, Shimokawa H (2017) Endothelial Functions. Arterioscler Thromb Vasc Biol 37: e108-e114.
- Barbosa AM, Silva KSF, Lagares MH (2017) Atherosclerosis: analysis of the eNOS (T786C) gene polymorphism. Genet Mol Res 16.
- Sung JH, Lee BE, Kim JO (2015) Association between eNOS polymorphisms and risk of coronary artery disease in a Korean population: a meta-analysis. Genet Mol Res 14: 16508-16520.
- Wang L, Li X, Zhang Y, Huang Y, Zhang Y, et al. (2018) Oxymatrine ameliorates diabetes-induced aortic endothelial dysfunction via the regulation of eNOS and NOX4. J Cell Biochem.
- Wang H, Qiu L, Ma Y (2017) Naoxintong inhibits myocardial infarction injury by VEGF/eNOS signaling-mediated neovascularization. J Ethnopharmacol 209: 13-23.
- McCrindle BW, Ose L, Marais AD (2003) Efficacy and safety of atorvastatin in children and adolescents with familial hypercholesterolemia or severe hyperlipidemia: a multicenter, randomized, placebo-controlled trial. J Pediatr 143: 74-80.
- Yang X, Li Y, Sun L (2017) NaoXinTong Enhances Atorvastatin-induced Plaque Stability While Ameliorating Atorvastatin-induced Hepatic Inflammation. J Cardiovasc Pharmacol 69: 55-64.
- Yang J, Yan R, Roy A, Xu D, Poisson J, et al. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12: 7-8.
- Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35: 3375-3382.
- Swendsen null, Wang null (1986) Replica Monte Carlo simulation of spin glasses. Phys Rev Lett 57: 2607-2609.
- Zhang Y, Skolnick J (2004) SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem 25: 865-871.
- Zhu X, Mitchell JC (2011) KFC2: a knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins 79: 2671-2683.
- Wan X, Liu P, Jin X (2018) Electrospun PCL/keratin/AuNPs mats with the catalytic generation of nitric oxide for potential of vascular tissue engineering. J Biomed Mater Res A.
- Wu D, Hu Q, Zhu D (2018) An Update on Hydrogen Sulfide and Nitric Oxide Interactions in the Cardiovascular System. Oxid Med Cell Longev 2018: 4579140.
- Liao Q, Huang YM, Fan W, Li C, Yang H (2016) Endothelial nitric oxide synthase deficiency influences normal cell cycle progression and apoptosis in trabecular meshwork cells. Int J Ophthalmol 9: 799-803.
- Kader KN, Akella R, Ziats NP (2000) eNOS-overexpressing endothelial cells inhibit platelet aggregation and smooth muscle cell proliferation in vitro. Tissue Eng 6: 241-251.
- Lim KH, Ancrile BB, Kashatus DF, Counter CM (2008) Tumour maintenance is mediated by eNOS. Nature 452: 646-649.
- Barbosa AM, Silva KSF, Lagares MH (2017) Atherosclerosis: analysis of the eNOS (T786C) gene polymorphism. Genet Mol Res 16.
- Campedelli FL, E Silva KSF, Rodrigues DA (2017) Polymorphism of the gene eNOS G894T (Glu298Asp) in symptomatic patients with atherosclerosis. Genet Mol Res 16.
- Staal FJT, van der Burg M, Wessels LFA (2003) DNA microarrays for comparison of gene expression profiles between diagnosis and relapse in precursor-B acute lymphoblastic leukemia: choice of technique and purification influence the identification of potential diagnostic markers. Leukemia 17: 1324-1332.
- Silva K (2018) Hot spots and single nucleotide polymorphisms on the interaction interface of RAD51 and p53 complex. J Tre Bio Res 1: 1-5.
- Tannous I, Santos T, de Curcio J (2018) Involvement of Protein-Protein Interactions of eNOS and Genetic Polymorphisms in Coronary Artery Disease. Int J Clin Cardiol Res 2: 67-71.
- de Curcio JS, Lima RM, Oliveira LN (2018) Structure-based design of TFF3-PAR2 inhibitor peptides as a promising new therapeutic approach for endometriosis patients. M J Gyne 3: 6.
- Achary MS, Reddy ABM, Chakrabarti S (2006) Disease-Causing Mutations in Proteins: Structural Analysis of the CYP1b1 Mutations Causing Primary Congenital Glaucoma in Humans. Biophys J 91: 4329-4339.
- Norel R, Sheinerman F, Petrey D, Honig B (2001) Electrostatic contributions to protein-protein interactions: Fast energetic filters for docking and their physical basis. Protein Sci 10: 2147-2161.
- Zhang Z, Witham S, Alexov E (2011) On the role of electrostatics on protein-protein interactions. Phys Biol 8: 035001.
- Li L, Jia Z, Peng Y (2017) Forces and Disease: Electrostatic force differences caused by mutations in kinesin motor domains can distinguish between disease-causing and non-disease-causing mutations. Scientific Reports 7: 8237.
- Li Y, Huang Y, Swaminathan CP, Smith-Gill SJ, Mariuzza RA (2005) Magnitude of the Hydrophobic Effect at Central versus Peripheral Sites in Protein-Protein Interfaces. Structure 13: 297-307.
- Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D (2012) Relationship between hot spot residues and ligand binding hot spots in protein-protein interfaces. J Chem Inf Model 52: 2236-2244.
- Dong JY, Qin LQ, Zhang Z (2011) Effect of oral L-arginine supplementation on blood pressure: a meta-analysis of randomized, double-blind, placebo-controlled trials. Am Heart J 162: 959-965.
- Feng J, Hu Y, Yi H (2013) Two Conserved Arginine Residues from the SK3 Potassium Channel Outer Vestibule Control Selectivity of Recognition by Scorpion Toxins. J Biol Chem 288: 12544-12553.
- Tesei G, Vazdar M, Jensen MR (2017) Self-association of a highly charged arginine-rich cell-penetrating peptide. PNAS 114: 11428-11433.
- Liao SM, Du QS, Meng JZ, Pang ZW, Huang RB (2013) The multiple roles of histidine in protein interactions. Chem Cent J 7: 44.
- Cauët E, Rooman M, Wintjens R, Liévin J, Biot C (2005) Histidine−Aromatic Interactions in Proteins and Protein-Ligand Complexes: Quantum Chemical Study of X-ray and Model Structures. J Chem Theory Comput 1: 472-483.
- Zheng P, Cao Y, Bu T, Straus SK, Li H (2011) Single Molecule Force Spectroscopy Reveals that Electrostatic Interactions Affect the Mechanical Stability of Proteins. Biophys J 100:1534-1541.
- Zhu J, Luther PW, Leng Q, Mixson AJ (2006) Synthetic Histidine-Rich Peptides Inhibit Candida Species and Other Fungi In Vitro: Role of Endocytosis and Treatment Implications. Antimicrob Agents Chemother 50: 2797-2805.
- Rhys NH, Soper AK, Dougan L (2012) The Hydrogen-Bonding Ability of the Amino Acid Glutamine Revealed by Neutron Diffraction Experiments. J Phys Chem B 116: 13308-13319.
- Law MJ, Linde ME, Chambers EJ (2006) The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res 34: 275-285.
- de Jesus AJ, Allen TW (2013) The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. Biochimica et Biophysica Acta (BBA) - Biomembranes 1828: 864-876.
- Samanta U, Chakrabarti P (2001) Assessing the role of tryptophan residues in the binding site Protein. Eng Des Sel 14: 7-15.
- Dill KA (1990) Dominant forces in protein folding. Biochemistry 29: 7133-7155.
- Chaturvedi D, Mahalakshmi R (2014) Juxtamembrane tryptophans have distinct roles in defining the OmpX barrel-micelle boundary and facilitating protein-micelle association. FEBS Lett 588: 4464-4471.
- Gupta A, Zadafiya P, Mahalakshmi R (2014) Differential contribution of tryptophans to the folding and stability of the attachment invasion locus transmembrane β-barrel from Yersinia pestis. Sci Rep 4: 6508.
- Pace CN, Fu H, Fryar KL (2011) Contribution of Hydrophobic Interactions to Protein Stability. J Mol Biol 408: 514-528.
- Yan C, Wu F, Jernigan RL, Dobbs D, Honavar V (2008) Characterization of Protein-Protein Interfaces. Protein J 27: 59-70.
- Roth CM, Neal BL, Lenhoff AM (1996) Van der Waals interactions involving proteins. Biophys J 70: 977-987.
- Roy S, Das TK (2015) Study of Interaction Between Tryptophan, Tyrosine, and Phenylalanine Separately with Silver Nanoparticles by Fluorescence Quenching Method. J Appl Spectrosc 82: 598-606.
- Word JM, Lovell SC, Richardson JS, Richardson DC (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285: 1735-1747.
- Yokota A, Tsumoto K, Shiroishi M, Nakanishi T, Kondo H, et al. (2010) Contribution of Asparagine Residues to the Stabilization of a Proteinaceous Antigen-Antibody Complex, HyHEL-10-Hen Egg White Lysozyme. J Biol Chem 285: 7686-7696.
- Yusufaly TI, Li Y, Singh G, Olson WK (2014) Arginine-Phosphate Salt Bridges Between Histones and DNA: Intermolecular Actuators that Control Nucleosome Architecture. J Chem Phys 141: 165102.
- Zhang Z, Liu L, Gomez Casal R (2016) Targeting cancer stem cells with p53 modulators. Oncotarget 7: 45079-45093.
- Krotova K, Hu H, Xia SL (2006) Peptides modified by myristoylation activate eNOS in endothelial cells through Akt phosphorylation Br J Pharmacol 148: 732-740.




