Type II diabetes mellitus (T2DM) is a chronic metabolic disorder characterized by insulin insensitivity, hyperglycemia, and immune dysregulation. Recent findings have shown that T2DM has a significant impact on the skeletal system, including the impairment of the fracture healing process which commonly leads to non-union. Throughout the process, heterotypic interactions between different immune cells are required for the recruitment and differentiation of osteogenic cells vital for fracture repair. The purpose of this study was to compare inflammatory gene expression induced in T2DM with those occurring during fracture repair with a specific focus on immune cell expression. Using publicly available RNA-seq datasets and Ingenuity Pathway Analysis (IPA), we compared gene expression profiles of human diabetic and non-diabetic data to gene expression profiles of mice post-fracture. IPA core analysis of diabetic vs. non-diabetic immune gene expression revealed top canonical pathways (p-value < 1.0 x 10-6) involved in the Th1 Activation Pathway, Granulocyte Adhesion and diapedesis, and IL-7 Signaling, had an average activation z-score of -2.373, thus exhibiting a predicted inhibition when compared to non-diabetic controls. Additionally, top upstream inflammatory regulators such as TNF-α, IL-1B, and IL-6 also exhibited an average 3.5 log-fold reduction in expression. When examining gene expression in normal fracture repair, previous upstream inflammatory regulators exhibit an average 2.1 log-fold increase. Our results suggest that during fracture repair, the early immune response required for recruitment of osteogenic cells and repair is impaired in T2DM signaling.
inflammation, type 2 diabetes, fracture, myeloid, ingenuity pathway analysis
T2DM: Type 2 Diabetes Mellitus; MSCs: Mesenchymal Stem Cells.
In type 2 diabetes mellitus (T2DM), the glucose homeostatic function of insulinis impaired which leads to negative physiological changes ] like failed bone healing. Inthe U.S. annually, 790,000 diabetic fractures do not heal properly each costing $30,000more in patient care Unfortunately, the connection between the immune system, T2DM,and fracture repair is stillunclear. After fracture, the acute inflammatory response recruits’ cells to supporthematoma formation, callus formation, and osteogenesis Recruited mesenchymal stem cells(MSCs) can differentiate into osteogenic cells required for the generation of the callus Thisstabilizes the fracture and acts as a scaffold for deposition of mature lamellar bone In diabetics,this process takes significantly longer. Hamon et al. observed that diabetic rats have increasedrates of non-union and lower bone mineral density at 12 weeks post-fracture when compared tonon-diabetics Roy et al. correlated these changes with declines in transcription factors suchas Dlx5 are inhibited under hyperglycemic conditions and required for osteogenicdifferentiation. This inhibition is thought to reduce MSC commitment, which depletes theavailable osteoprogenitor cells necessary for fracturerepair. Recent studies have shown that immune cells such as macrophages are also criticalfor fracture repair. Mouse macrophage ablation studies in fracture models show asignificant impairment in the fracture healing process These myeloid derived immune cellssecrete mediators which regulate processes like matrix synthesis and osteogenesis Similarimmune components are known to be dysregulated in TD2M Therefore, in this study weuse Ingenuity Pathway Analysis to compare human T2DM gene expression profiles with thoseof mice post-fracture to determine connections between immune dysregulation, T2DM, andfracture repair.
Literature search utilizing ingenuity pathway analysis
The Ingenuity Pathway Analysis (IPA, Qiagen, The Netherlands) Knowledge Basewas utilized to locate articles with RNA-seq and gene expression data related to diabetes,fracture repair, and myeloid cell activity. IPA’s BioProfiler function was used to search for specificgene functions and molecule activity while excluding drug and chemical markers. Theliterature search yielded only two studies, “Diabetic complications and dysregulated innate immunity”and “Transcriptomic signatures reveal immune dysregulation in human diabetic andidiopathic gastroparesis” that contained RNA-seq data that overlapped with our search criteriaand were used for subsequent meta-analysis and coreanalysis.
Comparison of gene expression in T2DM, non-diabetic, and acute fracture repair
First, the data were extrapolated and used to analyze transcriptome patterns ofhuman T2DM to non-diabetic and mouse non-diabetic unfractured to mouse non-diabeticpost-fracture with a specific focus on immune cell related gene expression andfunction. In Grover et al.’s study, gene expression data from smooth muscle wascompared between T2DM patients vs. non-diabetics controls Data were uploaded to IPA inExcel files and annotated prior to core analysis using IPA’s flexible file format. For data obtainedfrom Grover et al. 564 genes were inputted for analysis with 549 gene IDs successfully mappedand 15 gene IDs remaining unmapped and no additional cutoffs to fold-change in expression andp- values wereassigned.
In Bais et al.’s study, gene expression data was extracted at day 3 post-fracture, whichis established as part of the acute inflammatory phase of fracture repair in mice. For Bais etal. 16,505 genes were inputted for analysis with 13,454 gene IDs successfully mapped 3051gene IDs remaining unmapped. For this data set, cutoff values of -1.5 and +1.5 were implementedfor fold change in expression to reduce the analysis of genes inputted to 1998genes. Since both T2DM and fracture transcriptome represent sustained inflammatoryevents, these two datasets were compared to each other to find links between immunedysregulation, T2DM, and fracture repair that span both the mouse and humansystems.
Data IPA analysis
Predictions of the activation states of pathways and regulators was conductedthrough IPA’s z-scoring system. These z-scores act as a prediction model in which IPA canaccurately state if a specific pathway or molecule will be activated or inhibited based on geneexpression datasets uploaded to its core expression analysis feature. IPA can then compare theexpression patterns between these uploaded datasets and the existing datasets in the IPA KnowledgeBase. Any inconsistencies between the uploaded datasets and the IPA Knowledge Base willsubtract from the overall z-score while consistencies add to the z-score. A negative z-score isindicative of a predicted state of inhibition while a positive z-score represents a predicted stateof activation. When analyzing canonical pathways, the top 20 results (consisting of the lowest20 p-values) were selected for each category. For upstream regulators, the top 15 results(consisting of the lowest 15 p-values) wereselected.
Canonical signaling pathways involved in T2DM dysregulation of the immune response
When the uploaded dataset from Grover et al. was uploaded to the IPA, IPA predicted the top 20 canonical pathways, based on lowest p-values, to be inhibited overall (Figure 1, blue- shaded bars). Immune and inflammatory signaling pathways such as the Th1 pathway, IL-7 pathway, and Granulocyte Adhesion and Diapedesis functions all exhibited an average negative z-score of -2.737 (Figure 1, Table 1).
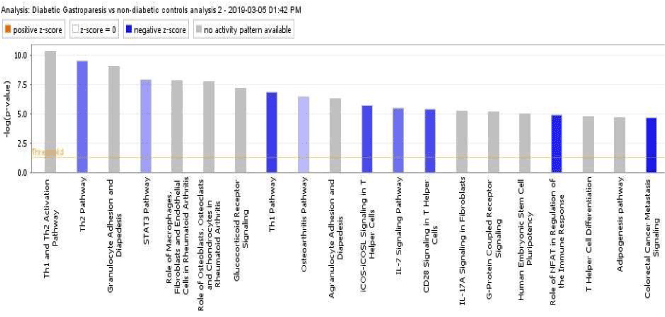
Figure 1. Canonical signaling pathways involved in T2DM dysregulation of the immune response
Table 1. Z-scores and P-values of Top 7 Downregulated Canonical Pathways in T2DM
Pathway |
Z-score value |
p-value |
TH2 Pathway |
-2.11 |
2.22E-10 |
Granulocyte Adhesion and Diapedesis |
-2.113 |
8.46E-09 |
STAT3 |
-1.414 |
1.31E-08 |
TH1 Pathway |
-2.887 |
1.46E-07 |
Osteoathritis Pathway |
-1 |
3.87E-07 |
iCOS-iCOSL Signaling T Helper Cells |
-2.714 |
1.79E-06 |
IL-7 Signaling Pathway |
-2.121 |
3.16E-06 |
Expression of Pro-inflammatory Mediators IL1-β and TNF-α are Significantly Inhibited in T2DM
In addition to top canonical pathways, IPA also predicted the activation states of similar upstream regulators using the same z-scoring system. In the T2DM dataset, inflammatory molecules TNF-α, IL-1β, and IL-6 were predicted to be inhibited with an average negative z-score of -5.952 (Table 1). Aside from cytokines, other endogenous inflammatory molecules such as prostaglandin E2 and leukotriene D4 were predicted to be inhibited with z-scores of -3.175 and -5.281, respectively (Table 2).
Table 2. Predicted activation states of inflammatory genes in type 2 diabetics
Upstream Regulator |
Molecule Type |
Predicted Activation State |
Activation z-score |
p-value of overlap |
TNF |
Cytokine |
Inhibited |
-6.036 |
9.84E-61 |
IL1β |
cytokine |
Inhibited |
-6.451 |
2.83E-46 |
beta-estradiol |
chemical - endogenous |
Inhibited |
-4.474 |
2.8E-40 |
PDGF BB |
complex |
Inhibited |
-7.094 |
6.1E-40 |
TGFB1 |
growth factor |
Inhibited |
-4.843 |
1.29E-39 |
prostaglandin E2 |
chemical - endogenous |
Inhibited |
-3.175 |
5.51E-39 |
IFNG |
cytokine |
Inhibited |
-4.492 |
1.5E-34 |
leukotriene D4 |
chemical - endogenous |
Inhibited |
-5.281 |
1.66E-34 |
GPER1 |
G-protein coupled receptor |
Inhibited |
-5.202 |
1.05E-33 |
NR3C1 |
nuclear receptor |
Inhibited |
-2.549 |
4.16E-33 |
IL2 |
cytokine |
Inhibited |
-4.739 |
1.52E-28 |
IGF1 |
growth factor |
Inhibited |
-4.599 |
1.56E-28 |
CD40LG |
cytokine |
Inhibited |
-4.404 |
2.23E-28 |
HGF |
growth factor |
Inhibited |
-4.95 |
2.44E-28 |
IL6 |
cytokine |
Inhibited |
-5.371 |
1.96E-25 |
Canonical signaling pathways involved in early fracture repair in non-diabetic conditions
Gene expression from early fracture repair in non-diabetic conditions were uploaded to IPA’s core expression analysis feature. The top 20 canonical pathways from this expression dataset reveal varied states of pathway inhibition (blue shaded bars) and ac Pathway and the LPS/IL-1 Mediated Inhibition of RXR Function with an average z-score of 2.745 (Figure 2, Supplemental Table 3). Clear bars are indicative of a z-score of 0 and thus have no difference in activity while gray shaded bars, such as Atherosclerosis signaling, indicate that no general prediction pattern was available from IPA’s z-scoring system.
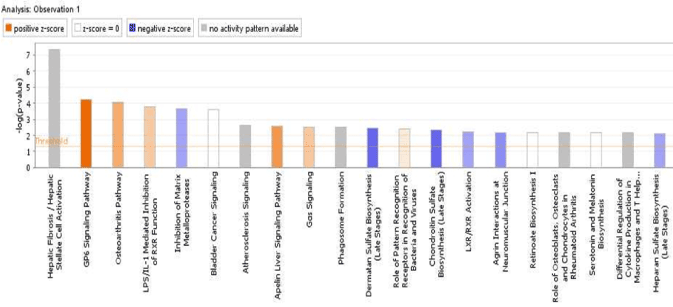
Figure 2. Canonical signaling pathways involved in early fracture repair in non-diabetic conditions
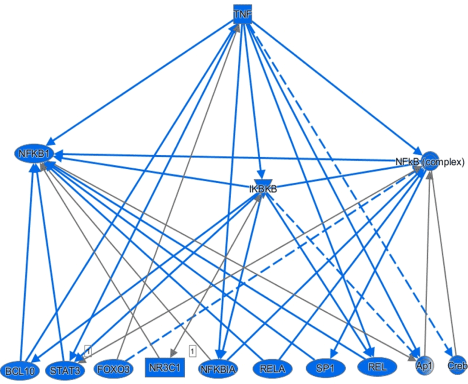
Figure 3. Predicted activity of downstream TNF-α signaling targets in type II diabetes
Table 3. Z-Scores and P-values for Top 7 regulated canonical pathways in non-diabetic fracture healing
| |
Z-score value |
p-value |
GP6 Signaling Pathway |
2.887 |
5.40E-05 |
Osetoarthritis Pathway |
1.604 |
8.93E-05 |
LPS/IL-1 Inhibiition of RXR Function |
3.783 |
1.65E-04 |
Inhibition of Matrix Metalloproteases |
-0.816 |
2.15E-04 |
Apelin Liver Signaling Pathway |
2 |
2.25E-03 |
Gαs Signaling |
1.134 |
-2.84E-03 |
Dermatan Sulfate Biosynthesis (Late Stage) |
-1.342 |
-3.57E-03 |
Table 4. Predicted activation states of inflammatory genes in non-diabetic fracture healing
Upstream Regulator |
Molecule Type |
Predicted Activation State |
Activation z-score |
p-value of overlap |
TGFB1 |
growth factor |
Activated |
5.235 |
91.94E-16 |
FGF2 |
growth factor |
Activated |
2.25 |
6.93E-14 |
BMP2 |
growth factor |
Activated |
3.484 |
1.71E-12 |
CD44 |
other |
Activated |
2.373 |
8.23E-11 |
TNF |
cytokine |
Activated |
2.908 |
2.11E-10 |
KRAS |
enzyme |
Inhibited |
-2.154 |
7.27E-10 |
SPP1 |
cytokine |
Activated |
2.174 |
1.88E-09 |
SMAD7 |
transcription regulator |
Inhibited |
-2.284 |
2.67E-09 |
RXRB |
ligand-dependent nuclear receptor |
Inhibited |
-2.236 |
3.12E-09 |
RUNX2 |
transcription regulator |
Activated |
2.391 |
5.48E-09 |
IGF1 |
growth factor |
Activated |
3.239 |
1.01E-08 |
P38 MAPK |
group |
Activated |
3.325 |
2.66E-08 |
Jnk |
group |
Activated |
3.413 |
3.05E-08 |
IL1β |
cytokine |
Activated |
3.311 |
3.09E-08 |
IL1α |
cytokine |
Activated |
4.221 |
8.15E-08 |
Expression of pro-inflammatory mediators IL1-β and TNF-α is upregulated early during fracture repair in non-diabetic conditions
Gene expression data sets pertaining to fracture repair in non-diabeticconditions uploaded to IPA’s core analysis predicted inflammatory molecules IL-1α, IL1-β, and TNF-αto be activated in early fracture repair with respective z-scores of 3.311, 4.221, and 2.908 (). Additionally, genes related directly to the initiation and progression of fracture repair, BMP2and RUNX2, were also predicted to be activated early during fracture repair innon-diabetic conditions (respective z-scores of 3.484 and2.391).
Inhibition of upstream inflammatory mediator TNF-α results in general suppression of downstream transcription factors involved in inflammation and enhancement of osteogenesis
IPA’s mechanistic network feature for reduced TNF-α activity in T2DM revealsan overall suppression of downstream signaling targets (). The signaling networksscores predicted activation states of downstream targets based on the z-scores of upstreamregulators seen in table 1. The downstream network shows that all downstream signaling are predictedto have suppressed or inhibited activity under hyperglycemic conditions. Some of thesetargets include transcription factors involved in inflammation (NF-kβ) and transcriptionfactors implicated in the enhancement of osteogenesis(STAT3).
The immune response plays a significant role in the process of fracture repair and itis often dysregulated in T2DM. These inflammatory responses are driven by immunesignaling pathways and the release of cytokines at the fracture site Our analysis shows asignificant dysregulation and suppression of canonical immune signaling pathways in T2DM includingthe Th1, granulocyte adhesion and diapedesis, and TNF-α regulation which all play a significantrole in host inflammatory responses (). The Th1 response requires high levels of inflammation and causes a strongcell-mediated immune response The Th1 response is important to macrophage activation, whichis essential to fracture repair We hypothesize that a reduction in Th1 signaling activityin T2DM could reduce available macrophages at the fracture site contributing to impairedfracture repair. Inflammatory cytokines TNF-α and IL-6 were also predicted to be inhibited inT2DM and activated post-fracture (). Interestingly, TNF-α and IL-6 are both secreted byTh1 cell subsets, which in turn activate macrophages Thus, a decrease in theseinflammatory cytokines could be related to the decrease in the Th1 pathway response and a reductionof macrophage activity at the fracture site. In addition to the Th1 pathway, thegranulocyte adhesion and diapedesis pathway is vital for myeloid cells such as macrophages andneutrophils to reach the fracture site and continue to promote this necessary inflammation. A reductionin this activity would result in reduced myeloid cell persistence at the fracture site, diminishingthe inflammatory response
Our data shows that in T2DM, inflammatory cytokines important for fracture repairare down regulated significantly (i.e. IL-1β and TNF, Comparing Table 1 to Table 2, z-score >-2). IL-1β functions during early fracture repair to recruit the necessary osteogenic cells andis consistent with IPA analysis of the post-fracture data set which shows activation ofIL-1β (). When IL1-β deficient mice were given IL-1β during fracture repair, fracturerepair improved when compared to controls Interestingly, other studies have shown IL-1βcan also be a potent inhibitor of chondrogenesis and an activator of osteogenesis In thissense, our findings could indicate that suppression of IL-1β in T2DM impairs the earlyinflammatory response and decreases osteoblast bonedeposition. TNF-α also plays a significant role in fracture healing by promoting MSCrecruitment and differentiation to osteoclast [22]. Data has shown that treatment with low doses ofTNF-α can enhance the rate of callus mineralization and fracture healing in rodent fracture modelsTranscription factors downstream of TNF-α such as NF-kβ and STAT3 were also predicted tobe suppressed in our signaling network (). NF-kβ and STAT3 activation are essentialfor osteoclasts and osteoblasts function During early fracture repair osteoclasts assist incallus reabsorption while osteoblasts deposition mature bone. STAT3 deficient mice afterfracture have impaired bone formation with reduced numbers of osteoblasts and increasedosteoclasts . It is hypothesized that a reduction of these mediators in T2DM could reduce theMSC number at the fracture site and negatively affect osteoprogenitor recruitment andfunction.
Overall, our data suggests that inflammatory pathways and molecules necessaryduring the early stages of fracture repair are suppressed in T2DM. Our IPA analysis providesinsight into which signaling pathways and transcription targets are important when elucidatingthe connection between the immune system, T2DM, and bonerepair.
- Eledo BO, Buseri F, Akhogba AO (2015) Evaluation of some haematological parameters among pregnant Ijaw Women: An indigenous West African tribe. J Health Med Nurs 1: 10-15.
- Azab EA, Albasha MO, Elhemady SY (2017) Haematological parameters in pregnant women attending antenatal care at Sabratha Teaching Hospital in Northwest, Libya. Am J Laboratory Med 1: 61-69.
- Boundless (2015) Introduction to pregnancy and human development. Boundless Anatomy and Physiology. Boston, MA: Boundless.
- Mohamed AO, Hamza KM, Babker AMA (2016) Physiological changes in some hematological and coagulation profiles among Sudanese healthy pregnant women. Int J Med Sci Public Health 5: 525-527.
- Chandra S, Tripathi AK, Mishra S, Amzarul M, Vaish AK (2012) Physiological Changes in Hematological Parameters during Pregnancy. Indian J Hematol Blood Transfus 28: 144-146. [Crossref]
- Canzoneri BJ, Lewis DF, Groome L, Wang Y (2009) Increased neutrophil numbers account for leukocytosis in women with Preeclampsia. Am J Perinatol 26: 729-732. [Crossref]
- Morelli S, Mandal M, Goldsmith LT, Kashani BN, Ponzio NM (2015) The maternal immune system during pregnancy and its influence on fetal development. Res Reports Biol 6: 171-189.
- Wallace AE, Fraser R, Cartwright JE (2012) Extravillous trophoblast and decidual natural killer cells: a remodelling partnership. Hum Reprod Update 18: 458-471. [Crossref]
- Nancy P, Erlebacher A (2014) T cell behavior at the maternal-fetal interface. Int J Dev Biol 58: 189-198. [Crossref]
- Surabhi AK Sanjay M (2012) Physiological Changes in hematological parameters during pregnancy. Indian J Hematol Blood Transfus 28: 144-146.
- Liu S, Diao L, Huang C, Li Y, Zeng Y, et al. (2017) The role of decidual immune cells on human pregnancy. J Reprod Immunol 124: 44-53. [Crossref]
- Dutta S, Sengupta P (2017) Defining pregnancy phases with cytokine shift. J Pregnancy Reprod 1: 124.
- Schumacher A, Costa SD, Zenclussen AC (2014) Endocrine factors modulating immune responses in pregnancy. Front Immunol 5: 196. [Crossref]
- Piccinni MP (2010) T cell tolerance towards the fetal allograft. J Reprod Immunol 85: 71-76. [Crossref]
- Sykes L, MacIntyre DA, Yap XJ, Teoh TG, Bennett PR (2012) The Th1:Th2 dichotomy of pregnancy and preterm labour. Mediators Inflamm: 967629. [Crossref]
- Williams PJ, Searle RF, Robson SC, Innes BA, Bulmer JN (2006) Decidual leucocyte populations in early to late gestation in normal human pregnancy. J Reprod Rev Immunol 6: 584e594. [Crossref]
- Aina O, Dadik J, Charurat M, Amangaman P, Gurumdi S, et al. (2005) Reference values of CD4 T-lymphocytes in human immunodeficiency virus-negative adult Nigerians. Clin Diagn Lab Immunol 12: 525-530. [Crossref]
- Audu RA, Idigbe EO, Akanmu AS, Mafe AG, Onyewuche J, et al. (2007) Values of CD4+ T lymphocyte in apparently healthy individuals in Lagos, Nigeria. Eur J Sci Res 16: 168-173.
- Oladepo DK, Idigbe EO, Audu RA, Inyang US, Imade GE, et al. (2009) Establishment of reference values of CD4 and CD8 lymphocyte subsets in healthy Nigerian adults. Clin Vaccine Immunol 16: 1374-1377. [Crossref]
- Akinbami AA, Dosunmu AO, Adediran A, Adewunmi AA, Rabiu KA, et al. (2014) Cluster of differentiation 4+ cell count mean value, reference range and its influencing factors in Human Immunodeficiency Virus-seronegative pregnant women in Lagos. Niger Med J 55: 116-120. [Crossref]
- Ramsay M (2018) Normal cellular changes during pregnancy and the puerperium. The Obstetric Hematology Manual. Cambridge University Press. Cambridge, UK.
- Opara EI, Zaidi J (2007) The interpretation and clinical application of the word 'parity': a survey. BJOG 14: 1295-1297. [Crossref]
- Hoffman W, Lakkis FG, Chalasani G (2016) B cells, antibodies and more. Clin J Am Soc Nephrol 11: 137-154. [Crossref]
- Abbassi-Ghanavati M, Greer LG, Cunningham FG (2009) Pregnancy and laboratory studies: a reference table for clinicians. Obstet Gynecol 114: 1326-1331. [Crossref]
- Chama CM, Morrupa JY, Abja UA, Kayode A (2009) Normal CD4 T-lymphocyte baseline in healthy HIV-negative pregnant women. J Obstet Gynaecol 29: 702-704. [Crossref]
- Ekwempu AI, Ekwempu CC, Ikeh E, Olabode A, Agaba E (2012) Comparison of CD4 cell counts in pregnant HIV-seropositive and HIV-seronegative Nigerian women. Lab Medi 43: 168-171.
- Babatope IO, Isabu PA, Imarenezor EPK, Adesanya TM, Ikimiukor AP (2018) Normal CD4, CD8 T-lymphocytes and leucocyte baseline in healthy HIV-seronegative pregnant women in Ekpoma, Edo state, Nigeria. Int J Basic Applied Innovative Res 7: 18-28.
- Kieffer TEC, Faas MM, Scherjon SA, Prins JR (2017) Pregnancy persistently affects memory T cell populations. J Reprod Immunol 119: 1-8. [Crossref]



