In this article, we aim to extract information from a large online clinical research database; demonstrate the pitfalls of data quality; and create a computer framework for the visualization of the results. Data were downloaded from the clinicaltrials.gov website and loaded into a relational database after cleaning. Network analysis methods were applied to find hierarchical relations and inspect temporal connections in the database. A web-based, open source application “h-Vis” (Hierarchical Data Visualization) was developed to visualize the results. Our results highlight some data quality issues and show what steps were necessary for the cleaning and analysis of the data. Despite the lack of uniform data quality, we were able to extract information from the database and present some statistical overview and visualization examples. Institution-level examination was impossible due to a lot of missing or inaccurate data. City-level connections between the research sites were extracted. Using the visualization tool, it is possible to identify cities with experience in certain disease or medical intervention, or to extract collaboration between medical centres. The extracted information can be useful for researchers to see the latest trends in medical research reaching clinical study phase. In addition, they provide a good orientation for the general audience (e.g. decision makers, investors) about the focus of clinical problems investigated in certain countries or cities. Our custom, publicly available framework might enable to perform further analysis on the data.
hierarchical data visualization, clinical trials
Transparency is very important demand against clinical trials, and it is commonly investigated in the recent medical literature [1-3]. Public data repositories are essential tools for fulfilling this requirement: they can provide information to every interested actor, including health professionals, policy makers, health authorities and the general public [2]. Thanks to information technology solutions and advanced Internet-based services, these repositories can be easily accessed by anyone. The first registries were created as an answer to the publication bias discovered in the 1980s [4]. In the early 2000s dozens of small registries existed parallel, operated by among others hospitals and academic centres or pharmaceutical companies [5]. Many governments have recognized the need for national or even international level clinical trial databases. As a result, a number of systems were developed including the European Clinical Trials Database (EudraCT), the Australian New Zealand Clinical Trials Registry (ANZCTR), the Japan Primary Registries Network (JPRN) etc., [4]. The World Health Organization has developed a common platform called the International Clinical Trials Registry Platform (ICTRP) which regularly imports data from 17 sources, providing a single access point to a common, core dataset [6].
The 1997 Food and Drug Administration Modernization ACT (FDAMA) required to maintain a public database of the clinical trials performed in the US [7]. As a result, the National Library of Medicine has developed the clinicaltrials.gov website which is available since 29. February 2000 [8]. There are several regulations and policies prescribing mandatory data upload for certain types of clinical studies, but voluntary data supply is also available, including studies performed outside the US [3,9]. As a result, the database has grown to the largest world-wide database of medical research activities.
Most of the studies are clinical trials, but observational studies are allowed as well [10]. It is possible, and for certain types of studies, it is mandatory to provide data regarding the study result [10]. The data are uploaded by the sponsor or principal investigator of the study through a web-based form [10].
Recently many complex biological, social or communicational systems were described and analysed using network theory and methods [11]. In these models, the individuals, organizations or devices are represented by the nodes of the graph, and the edges (links) show the connections between them [12]. The most commonly studied networks include among others the Internet, the social networks and epidemiological applications [13,14]. These models and methods can be applied to clinical trials data collected in large relational databases in order to get a general overview of the domain and to explore connections such as temporal [15] or hierarchical relationships within the data [16].
Network visualization methods were used among others to assess the collaboration of scientific institutes in a geographical area [17]; to find potential collaborators or to support interdisciplinary innovation [18,19]. The most commonly used source of collaboration information is the co-authorship of publications. However, this approach is not suitable if we would like to explore the collaboration network of clinical studies: some of the results may not be published; ongoing and planned studies are not yet published; or some of the collaborators may not be mentioned as authors [20-22]. When designing a new clinical study, the selection of the study sites could have a significant impact on the success [23,24]. Besides the geographical diversity, it is worth to consider the “disease-profile” of the potential sites - former studies in similar topics could predict the presence of suitable patient population and knowledge necessary for a successful study. Recent research on the secondary use of clinical trials database content was focused on the eligibility criteria features [25,26] or the target population [27]. Another paper presented a method for map-based visualization of clinical study locations for a selected disease [28].
In this paper, our first aim is to investigate the data quality and general characteristics of the information available from clinicaltrials.gov database. In addition, we aim to provide tools to aid investigation of the data set. We present some examples of information extraction and visualization based on methods from network analysis applied to this data set. We develop a computer framework which supports performing such research within the data set. As opposed to previous research, we focus on finding connections between the study sites in order to build a trial network. Our results may support the design of new clinical studies as well as obtaining additional information from former research.
The content of the clinicaltrials.gov database is freely available for download and further analysis as separate XML files per study. In order to facilitate the interpretation of the data, a custom JAVA software was developed for pre-processing the data. The XML files were processed one by one, and the extracted data elements were loaded into a relational database implemented in MySQL. (Authors note: After the work described in this article was performed, the Clinical Trials Transformation Initiative made available an up-to-date relational database version of the database, hence now this first step could have been omitted.)
Clinicaltrials.gov is a large multi-source database which lacks uniform data quality, especially when the upload form allows entering free-text data. This might be sufficient for the analysis and control of elementary data, but does not allow large-scale comparisons [29]. Common problems in such datasets include among others missing values, misspellings, embedded values (multiple values entered in one field), misfielded values (value is entered not in the right field) [30]. Therefore data cleaning is an essential step before extracting information from the database [30].
The database contains information about the research locations of the studies. One study might have multiple locations, and for each of them, separate XML tags for the institution name, country, city and address are available, but all of these are free-text field thus do not have a uniform value set. Therefore, data cleaning was necessary before any further analysis. It was first performed on the city names using the open-source software OpenRefine [31]. After applying the built-in clustering algorithms, additional manual review was used to achieve maximal cleanliness. If the city was unidentifiable, the name was set to NULL and the location was excluded from further analysis. Otherwise, the most commonly used variation was selected.
The quality and data cleaning possibilities of institution names was also inspected. We have found that in many cases, only an ID-number or the name of the pharmaceutical company performing the study was given as institution name. A pilot analysis was made on the Hungarian study locations, and it was found that only less than 20 per cent of the institution names were identifiable even after extensive manual data cleaning. So, it was concluded that the data quality of the database does not allow institution-level analysis, only city-level.
Besides the location, the second target of our analysis was the targeted disease or the performed intervention (e.g. medication used) of the clinical studies. In the XML schema of the database there are three data fields (tags) which might provide information about these data:
- Condition and Intervention tags
- Keywords
- MeSH (Medical Subject Headings) terms
The data quality of all the three options was investigated (see Results section for examples of the issues found) and the MeSH terms were selected for further analysis.
Descriptive statistics were created using SQL queries and Microsoft Excel. Extraction of some parameters was performed using custom JAVA codes. Connections between the research locations were analysed by applying the following algorithm to each MeSH term, implemented in JAVA:
- A database query was performed in order to list the cities where at least one study with the given term took place. The cities were ordered by the date of the first occurrence of the term. The resulting cities are selected as the nodes of the constructed graph.
- The algorithm iterated through the cities in a reverse order, and for each of them, it has identified the city which had the earliest common study with the given one. An edge was created between these two cities.
Many studies have multiple associated conditions or interventions, so it was possible to build a hierarchy based on the co-occurrence of the terms using the method described in [16].
A web-based tool “h-Vis” (Hierarchical Data Visualization Tool) was developed to provide a visual overview of the extracted data. The visualization was created with open-source JavaScript libraries (d3.js and dhtmlXGrid) with a PHP backend for some functions. The open-source software code is available from https://gitlab.com/ttamas85/h-vis and a running prototype is accessible at http://ujrr.sote.hu/h-vis/.
The main functions of the tool include:
- Visualization of the locations (cities) of studies tagged with a selected MeSH term
- Visualization of the MeSH hierarchy built on co-occurrence
- Visualization of the locations (cities) of studies tagged with at least one of the selected MeSH terms
- Map-based visualization by country or region
Data quality and data cleaning
In this section we provide examples of data quality issues experienced during the selection of data elements suitable for further analysis. First, we discuss the medical targets (disease and intervention) of the studies, and after that the geological locations.
The dedicated fields named “Condition” and “Intervention” are both free-text fields thus proved to have a poor data quality. Typical problems include the following:
- multiple values are given in a comma separated list;
- longer, complex expressions are entered (e.g. “Child or Adolescent Bipolar I Disorder, Manic or Mixed Episode with or without Psychotic Features”);
- the most common issue is the use of synonyms and language variations (e.g.: Diabetes Mellitus Type 2; Diabetes Mellitus, Non-Insulin-Dependent; Diabetes Mellitus, Type 2; Diabetes Mellitus, Type II; Diabetes, Type 2; Type 2 Diabetes; Type 2 Diabetes Mellitus; Type II Diabetes Mellitus);
- in case of intervention, the dose of the medication is often included in the field value.
The values of the “keyword” field were also checked, but it has similar and even more severe quality issues. For example, a comma separated list of keywords was often provided instead of creating a separate tag for each keyword as expected. The comma was not always used as a separator (e.g. values like "pneumonia, bacterial", "Transplantation, kidney", "Transplantation, renal" are common), therefore it was not possible to split the values automatically. In some cases, whole sentences describing the study were entered as a keyword.
Due to these quality issues, the use of MeSH (Medical Subject Headings) terms assigned to the studies were considered for further analysis. The only drawback of using these data elements is that they are selected by an algorithm, not by the uploader: a weighted search is performed on the data for the MeSH terms and their synonyms (source: email communication with the clinicaltrials.gov customer support). As the MeSH is a controlled vocabulary, it lacks the issues of the free text data items. Despite of the possible errors of the automated assignment, still the MeSH terms seemed to be the best candidate for further analysis without extensive manual data cleaning. The terms are categorized into condition terms and intervention terms.
For city names, the following major data quality issues were identified:
- non-interpretable values which do not refer to a particular city, for example:
- “multiple cities” / “many cities”
- country name
- cities in (country name)
- “TBD”, “unknown”
- street address given in the city field
- name variations:
- English and native name variants
- various abbreviations like St/St./S/Saint
- inclusion of district/city part (with or sometimes without the city name)
- transcription variations of non-English characters
- typos
Most of these issues were manageable by performing data cleaning on the values of the “city” field, however it required significant manual and semi-manual effort.
Overview of the data set
The complete clinicaltrials.gov database was downloaded for analysis which resulted 194792 studies from the year range 1966-2020 (date of download: 29. 07. 2016). The MeSH terms are assigned in two categories: condition terms and intervention terms. 6447 different MeSH terms appeared in our data set. In average 2.06 condition terms and 2.25 intervention terms are assigned per study, the highest number was 62 conditions and 118 interventions, but only a few studies have more than 10 terms assigned.
The database contains studies from 191 different countries, hence most of the world. As expected, the United States has the most studies, but other countries also have a remarkable quantity. 89% of the studies are performed in a single country, 99% have no more than 15 countries while the highest number is 60 and the average is 1.56.
The studies have altogether more than 1.6 million locations (one site of a study is considered as a location, so this is not the number of unique research institutions). The average is 8.6 location per study (range: 1 to 3511), and 66% of the studies have a single location, although this ratio is highly variable among the countries (Table 1).
Table 1. Countries with the most studies included in the clinicaltrials.gov database
Country |
Study count |
Location count |
Single location studies |
1. United States |
93967 |
775660 |
57184 |
61% |
2. Canada |
15506 |
56603 |
6427 |
41% |
3. France |
14519 |
96185 |
5847 |
40% |
4. Germany |
14481 |
104914 |
4947 |
34% |
5. United Kingdom |
12098 |
46591 |
4584 |
38% |
6. Italy |
8790 |
44156 |
2512 |
29% |
7. Spain |
8425 |
43624 |
1967 |
23% |
8. China |
7853 |
26717 |
5558 |
71% |
9. Korea, Republic of |
7234 |
21845 |
4140 |
57% |
10. Netherlands |
6833 |
20269 |
2251 |
33% |
Total |
194792 |
1648260 |
129470 |
66% |
The extraction algorithm identified 6412 MeSH terms. This is slightly less than the number of terms described in the previous section as the studies associated with some terms have either no location or no start date stored in the database and these studies were excluded by the algorithm.
In this section, some parameters of the resulting graphs are presented. The average count of the cities (nodes) is 233 per MeSH term; the intervention terms have a slightly higher city count than the condition terms (243 vs. 193). 736 terms occurred in only one city while the highest number of cities is 8384 for the term “Hypertension”. The following ten terms have the most cities:
- Hypertension
- Diabetes Mellitus, Type 2
- Diabetes Mellitus
- Pulmonary Disease, Chronic Obstructive
- Asthma
- Lung Diseases
- Gastroesophageal Reflux
- Anti-Inflammatory Agents, Non-Steroidal
- Atrial Fibrillation
- Syndrome
Most of these are terms describing conditions, mainly chronic diseases. Almost each of them is very general category, so the list of the most frequent terms can only be used to get an insight about the focus points of medical research, but these terms are not suitable for detailed analysis. There is only one intervention term in this list, which is a very general and commonly used group of drugs (Anti-Inflammatory Agents, Non-Steroidal). The first particular agent is an anti-hypertension medicine: Telmisartan (18th highest number of cities, 4370).
Some of the cities are not connected with edges to other cities. This means that they have not had a common study with any other city with the inspected MeSH term. The number and ratio of these “orphan” cities were inspected. The results showed that the average count of the orphan nodes is 16 per term. Most terms have an orphan ratio less than 10%. A few terms do not have any links between the cities (i.e. orphan ratio is 100%), but these have a small number of cities (usually less than 10).
For the majority of the terms (4448 - 69.4%), the resulting graph had exactly one connected segment (and eventually some orphan nodes). 795 terms (12.4%) had no connection at all. The remaining had two or more segments. There was no significant difference between the condition and intervention terms. The average segment count is 1.4.
Another parameter which shows the characteristics of the resulting graphs is the degree of the nodes which is defined as the number of edges the node has to other nodes. As our graph is directed, nodes have two different degrees, the in-degree, which is the number of incoming edges, and the out-degree, which is the number of outgoing edges. In our case, the in-degree is always 1 (or 0 in case of the orphan nodes). Most terms have a rather small average degree, only 104 terms (1.6%) are greater than 10.
Visualization tool
A small web application was developed to provide a visual overview of the results. The MeSH terms are listed in a sortable and filterable table, showing some basic characteristics like the number of nodes, the start date of the first and last study etc. Two types of visualizations were developed:
- Graph: providing an overview of all nodes of the selected term. The cities are depicted as color-coded circles (yellow: the earliest location; red: the latest location) connected by arrows. It uses a force-directed layout. Details like city and country name or exact start date can be displayed on mouse over.
- Tree: displays details of a selected fragment of the graph as a tree. The city and country names are written next to the node. The same colour scale is used as in the graph. Branches of the tree can be collapsed and expanded.
As the generation of the data structures for the visualizations required up to several minutes per MeSH term, a pre-processing was performed: a JAVA code was developed to process each term and store the results as JSON files. The visualization tool uses these files as data sources.
Figures 1 and 2 depict the example of the rare disease “Marfan syndrome”. From the diagram we can identify the cities where this condition was investigated: most of them are in the US, there are some Western European cities (from the UK, Belgium, Denmark, France etc.) and a few from Eastern Asia. We can see among others that there is no reported clinical study from the Central and Eastern Europe region. From the graphs we can confirm that the research has started with single location studies followed by a large multi-centre study where most of the locations have former cooperation with institutes in Baltimore.
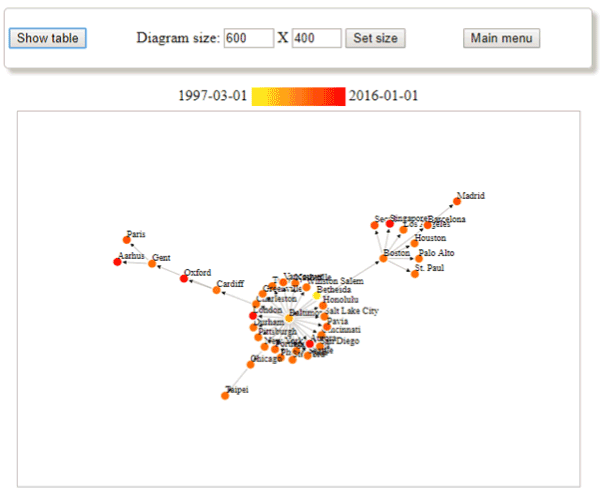
Figure 1. Example output of graph style visualization from the h-Vis tool, showing the extracted connections between the cities having studies in the clinicaltrials.gov database with the selected MeSH term “Marfan syndrome”
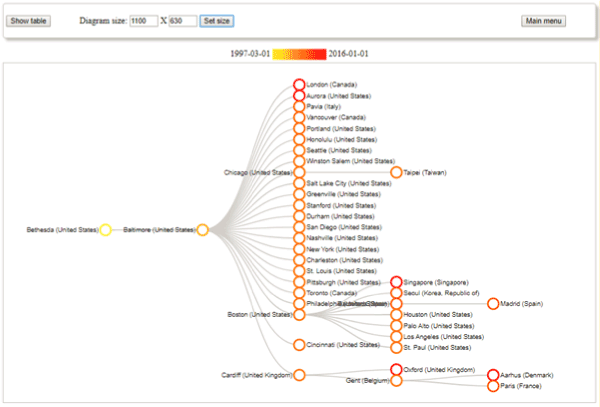
Figure 2. Example output of tree style visualization from the h-Vis tool, showing the extracted connections between the cities having studies in the clinicaltrials.gov database with the selected MeSH term “Marfan syndrome”
The hierarchy construction algorithm identified 3412 conditions and 2580 interventions as part of the hierarchies. As this means a very large tree, a visualization tool was developed which allows the selection of a partial tree. The user can select a MeSH term and define how many parent and/or child levels should be displayed. The sub-tree is dynamically calculated from the data. An example is shown in Figure 2. From these data, it is possible to identify the real connections between the terms. The constructed hierarchy often shows general-special term connections, but relationships between diseases could also be found.
The user can select one or more nodes in this tree and generate a combined graph or tree visualization. This means that all the cities hosting a research with any of the selected terms are listed and the algorithm is applied to this data set. This function is useful if someone is interested in locations of a general disease category as it cannot be guaranteed that the general term is associated with every study dealing with a special sub-type.
During the more than 15 years of its history, the clinicaltrials.gov database has become one of the largest sources of knowledge about clinical studies in the world which contains data from almost 200 thousand research projects from several decades. Despite its US origin, many studies performed outside the US are uploaded: it contains a large number of records among others from Europe or East Asia. It is interesting that these are not just research performed together with US sites, as for example 71 per cent of the Chinese records have a unique location. However, some free-text data fields cause data quality issues and make the interpretation and analysis of the data difficult. For example, a significant amount of data cleaning was necessary in order to achieve city-level analysis, but for institution-level evaluation even more effort would have been necessary. Standardizing the values of these fields could significantly improve the usability of the database. In addition, many location names include only an identification number or code used by the investigator which makes impossible to identify the given institution. Hence, some study coordinators (primarily big pharmaceutical companies) make these data private which is contradictory to the original goals of the database, and do not serve the transparency of clinical research.
The data available from the repository is sufficient for the analysis of individual studies or calculate descriptive statistical parameters, but if we aim to get a global overview and large-scale analysis, different methods are necessary. Restructuring the data in the form of networks and applying algorithms and visualization techniques from network theory can be sufficient for such goals. These require the accurate linking of data which cannot be achieved without cleaning the data. During this process a number of weak points were brought to the surface. These lessons learnt could be used for improving the existing database and for facilitating the design of new ones.
Our study showed through two examples that it is possible to analyse the information from the clinicatrials.gov database using network applications. However, it required extensive data preparation and data cleaning steps which could be achieved using both existing tools and custom software, but significant manual efforts were needed. Based on the MeSH terms describing the conditions and interventions related to the studies and the city names of the study locations it was possible to draw a “map” of the connections between the research sites. In addition, we have re-created the MeSH hierarchy based on real life co-occurrence of the terms.
Using the visualization tool, we can easily find solution to problems like these:
- Identify cities with experience in certain disease or medical intervention, particularly useful for rare diseases or new interventions;
- Find patients who were treated with certain medications in a given time frame, in order to perform long term follow-up studies;
- Identify local or international collaborations and relationships between medical centres.
Answering such questions is much more difficult using only the original website of the database.
Global, open-access databases like the clinicaltrials.gov are valuable source of information about past and current medical research. We have analysed this database from a new point of view: processing these data enables us to find trends, relationships and hierarchical connections within the data. However, as we have shown in the article, this kind of analysis requires extensive pre-processing and data cleaning, as the data quality is not uniform throughout the database. Our results highlight some data quality issues and show what steps were necessary for the analysis of the data. The lessons learnt might help in the improvement of the database or the design of further multi-source data collections. The issues make the reproducibility and the traceability of the clinical trials more difficult.
Despite the data quality issues, we were able to extract information from the database and present some statistical overview and visualization examples. Our custom, publicly available framework might enable to perform further analysis. The extracted information can be useful for researchers to see the latest trends in medical research reaching clinical study phase. In addition, they provide a good orientation for the general audience (e.g. decision makers, investors) about the focus of clinical problems investigated in certain countries, cities or in form of global cooperation. Improving the data validation methods of the database, like the substitution of free text fields with selection lists could enhance the data quality and enable more fine-grained analysis.
This research provides only a snapshot of the database. Automatic update is not possible due to the need of data cleaning. The main limitation of the model is that it works only on city level. For example, particularly in bigger cities there can be independent institutions or research groups which do not work together thus representing the city as a single node is not accurate enough. But from the currently available data it is not possible to perform a more fine-grained analysis as (i) cleaning of the institution names would require even more manual work and (ii) many location names are masked by codes and identifiers and it is impossible to reveal the exact facility.
- Chan AW, Tetzlaff JM, Altman DG, Laupacis A, Gotzsche PC, et al. (2013) SPIRIT 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med 158: 200-207.
- Dal-Re R (2015) Improving transparency of clinical trials. Trends Pharmacol Sci 36: 323-325.
- Hudson KL, Lauer MS, Collins FS (2016) Toward a new era of trust and transparency in clinical trials. JAMA 316: 1353-1354. [Crossref]
- Dickersin K, Rennie D (2012) The evolution of trial registries and their use to assess the clinical trial enterprise. JAMA 307: 1861-1864. [Crossref]
- Dickersin K, Rennie D (2003) Registering clinical trials. JAMA 290: 516-523. [Crossref]
- WHO (2019) International clinical trials registry platform (ICTRP) data provider.
- FDAMA (1997) Food and drug administration modernization Act of 1997, Pub. L. No. 105-115 Stat. 2310.
- NIHL (2000) National institutes of health launches clinicaltrials.gov.
- NIH (2016) Clinical trials registration and results information submission 2016.
- Zarin DA, Tse T, Williams RJ, Carr S (2016) trial reporting in clinicaltrials.gov - The final rule. N Engl J Med 375: 1998-2004. [Crossref]
- Amaral LA, Scala A, Barthelemy M, Stanley HE (2000) Classes of small-world networks. Proc Natl Acad Sci U S A 97: 11149-11152. [Crossref]
- Pastor-Satorras R, Vespignani A (2001) Epidemic spreading in scale-free networks. Phys Rev Lett 86: 3200-3203. [Crossref]
- Steitz BD, Levy MA (2016) A social network analysis of cancer provider collaboration. AMIA Annu Symp Proc 2016: 1987-1996.
- Benchimol EI, Bernstein CN (2017) Trends in epidemiology of pediatric inflammatory bowel disease in canada: distributed network analysis of multiple population-based provincial health administrative databases. Am J Gastroenterol 112: 1120-1134. [Crossref]
- Kasprzak R (2012) Diffusion in networks. Journal of Telecommunications and Information Technology 2012: 99-106.
- Tibély G, Pollner P, Vicsek T, Palla G (2013) Extracting tag hierarchies. PLoS One 8: e84133. [Crossref]
- Fung HN, Wong CY (2017) Scientific collaboration in indigenous knowledge in context: Insights from publication and co-publication network analysis. Technological Forecasting and Social Change 117: 57-69.
- Luong NT, Nguyen TT, Hwang D, Lee CH, Jung JJ (2015) Similarity-based complex publication network analytics for recommending potential collaborations. J UCS 21: 871-889.
- Schaar AK, Valdez AC, Ziefle M (2013) Publication network visualization as an approach for interdisciplinary innovation management. Professional Communication Conference (IPCC). IEEE International.
- Sterling TD, Rosenbaum WL, Weinkam JJ (1995) Publication decisions revisited: The effect of the outcome of statistical tests on the decision to publish and vice versa. The American Statistician. 49: 108-112.
- Dickersin K (1997) How important is publication bias? A synthesis of available data. AIDS Educ Prev 9: 15-21. [Crossref]
- Scherer RW, Langenberg P, Elm E (2005) Full publication of results initially presented in abstracts. The Cochrane Library.
- Gheorghiade M, Vaduganathan M, Greene SJ, Mentz RJ, Adams KF, et al. (2014) Site selection in global clinical trials in patients hospitalized for heart failure: perceived problems and potential solutions. Heart failure reviews 19: 135-152.
- Sarwar CM, Vaduganathan M, Butler J (2017) Impact of site selection and study conduct on outcomes in global clinical trials. Curr Heart Fail Rep 14: 203-209.
- Hao T, Rusanov A, Boland MR, Weng C (2014) Clustering clinical trials with similar eligibility criteria features. J Biomed Inform 52: 112-120.
- Miotto R, Jiang S, Weng C (2013) eTACTS: A method for dynamically filtering clinical trial search results. J Biomed Inform 46: 1060-1067.
- He Z, Carini S, Hao T, Sim I, Weng C (2014) A method for analyzing commonalities in clinical trial target populations. AMIA Annu Symp Proc 2014: 1777-1786.
- Luo J, Chen W, Wu M, Weng C (2017) Systematic data ingratiation of clinical trial recruitment locations for geographic-based query and visualization. Int J Med Inform 108: 85-91. [Crossref]
- Doan A, Ramakrishnan R, Vaithyanathan S (2006) Managing information extraction: state of the art and research directions. Proceedings of the 2006 ACM SIGMOD international conference on Management of data.
- Do ERHH (2000) Data cleaning: Problems and current approaches. IEEE Data Engineering Bulletin 23: 3-13.
- Ham K (2013) OpenRefine (version 2.5). http://openrefine.org. Free, open-source tool for cleaning and transforming data. J Med Libr Assoc 101: 233-234.


