In Human, Odors are detected by a large family of Olfactory Receptors (ORs) proteins. A sequence of Human ORs database has been proposed by D. Lancet et al. (2000, 2006) based on divergence of evolutionary model. In our earlier work, we have reckoned many important features (viz. Fractal dimension of Indicator matrix, DNA walk, Hurst exponent of 2-adic and 4-adic representations, codon compositions) of DNA sequences and their corresponding amino acid sequences of human ORs and also after making local changes we have calculated the same and found significant quantitative variations between the original and edited sequences. But it is a fact that two same sequences may function different and two different sequences may function same due their specific protein structure. So for complete understanding the effect of mutation we need to study the structure of proteins of those DNA sequences. In this article, an effort has been made to understand the effect of local mutations in the amino acid sequences as well as their tertiary protein structures of Human ORs. The significant changes are resulted through the present analysis through Fractals and standard Bioinformatics tools.
human olfactory receptors (ORs), protein sequences, fractal dimension & protein structure
ORs is the basis for the sense of smell, and they constitute the largest gene super family in the Human genome [1]. In Human genome, there are about 25000 protein-coding genes and out of which only 700 approximately ORs present [2,3]. However, these genes are more complex, with more alternative splicing generation of larger number of protein products.
Mutation is a biological process which are essentially an arbitrary insertion/ deletion/ replacement in the genomic DNA sequences. Effect of mutations occur in enzymes that do not function, abnormal protein structure or changes in protein conformation that result in a non-functioning or differently functioning protein.
In our earlier work, we have taken a class of human ORs sequences and their mutated sequences which are analyzed using Fractal we have been able to understand effect of edition (mutation) in DNA level. Some of the sequences are entirely been tracked out as per fractal observations and some of them still in the ranges. But through these analysis what we have lost to conclude is that whether they are at all functioning or malfunctioning. Essentially only syntactical changes in DNA are being encountered. So for complete understanding the effect of mutation we need to study the structure of proteins of those DNA sequences.
In this article, we have deciphered a quantitative details of effect of local mutation in protein through fractal analysis and standard Bioinformatics1.
Point mutations in a protein sequence may result in a change or loss of the native structure, which in turn may cause a change or loss of function [4], and ultimately yields different phenotypes. In addition to the natural variations among the individuals, researchers frequently introduce single amino acid residue replacements by misdirected mutagenesis in the laboratory to explore structural and functional features of proteins.
Experimental exploration of different positions in a protein structure with various residue types is a time consuming and expensive process. Such an exploration is generally facilitated by three dimensional (3D) modeling of side-chain mutations [5-9]. While the modeling of a single side chain in a given atomic environment seems to be one of the easiest of all protein structure prediction problems, it is still unsolved [10]. Seemingly insignificant change of a side-chain may lead to a significant change or loss of protein function [11]. This observation implies that side-chain conformation prediction is useful only if it is highly accurate, which makes it a challenging problem. Two simplifications are frequently applied in the modeling of side-chain conformations. First, amino acid residue replacements often leave the backbone conformation almost unchanged [12]. As a consequence, many algorithms x the backbone during the search for the best side-chain conformations. Second, it was observed that most side-chains in high-resolution crystallographic structures can be represented by a limited number of conformers that comply with stereo chemical and energetic constraints [13]. This observation motivated Ponder and Richards to develop the first library of side-chain rotamers for the 17 types of residues with dihedral angle degrees of freedom in their side-chains, based on 10 high-resolution protein structures determined by X-ray crystallography [14].
1 Data Used: We have taken a list of OR sequences of namely OR13G1, OR10K1, OR10T2, ,OR10R2, OR6Y1, OR10K2, OR2T1, OR2T4, OR2T5, OR2T6, OR6P1, OR10Z1, OR6K2, OR6K3, OR6K6, OR6N1, OR2L2, OR6N2, OR2L8, OR5AX1, OR5AY1, OR2G2, OR6F1, OR2G3, OR5AV10, OR11L1, OR10J1, OR2T3, OR1C1 and OR10J5 from OR database .
However, it seems difficult to directly apply traditional pattern matching algorithms [15-17] to 3D protein structure data. There are several qualitative bio-physical/chemical techniques through which we can analyze protein structures [18].
There are several methods to analyse and to compare tertiary protein structures. A method for matching curves that accommodates large and small deformations was implemented by Karp and Rabin [19]. This method preserves geometric similarities in the case of small deformation, and decreases geometric constraints when large deformations occur.
The approach is based on the computation of a set of geodesic paths connecting the curves. These two curves are defined as a source area and a destination area that can have an arbitrary number of connected components and different topologies.
In the present work, in understanding local mutation effect(s) on protein structures and functions, fractal geometric techniques have been employed [20]. We took a list of human olfactory receptors amino acid sequences and corresponding protein structures for this study. Before entering into the main work. Let us have a very brief look into fractals.
Our artificial world can be described easily through Euclidean geometric shapes. But there are many things in nature. Such as shape of cloud, geometry of lightening etc. could not be described through Euclidean geometry [21]. Many mathematicians descended the challenge for a fair enough description of natural objects. But after a long period in 1975, B. Mandelbrot took the challenge and gave the birth of a new geometry to describe nature which is known to us as Fractal Geometry in short Fractal. The precise definition of "Fractal" according to Benoit Mandelbrot is as a set for which the Hausdroff Besicovitch dimension strictly exceeds the topological dimension. To gain a quantitative insight of Fractal, some fractal parameters, namely Fractal dimension, Hurst exponent, succolarity, lacunarity etc. is also introduced in the literature. A brief discussion follows about one of the well-known methods of calculating fractal dimension, namely Box-Counting method [22].
This method computes the number of cells required to entirely cover an object, with grids of cells of varying size. Practically, this is performed by superimposing regular grids over an object and by counting the number of occupied cells. The logarithm of N(r), the number of occupied cells, versus the logarithm of 1/r, where r is the size of one cell, gives a line whose gradient corresponds to the box dimension.
Here we took thirty two protein sequences of Human ORs and their corresponding tertiary protein structures through Protein Structure Prediction Server ((PS)2) server [23].
Methods
After DNA sequence taken from ORDB [2] we have calculated the number of A,T,C and G occurrence in a single DNA sequence which has given in Tables 2 and 3, then we have edited the sequence to get a new sequence, after finding the position of each sequence. Before and after local editing in the sequences the detail account of frequencies of singletone nucleotide and polytone nucleotides is given in the Table 1. Using Bioinformatics toolbox of Matlab, we have determined the some of the graphs of fundamental protein properties, namely % of accessible residues (AR), Buried resides (BR), Alpha helix (Chou & Fasman) (AH), Amino acid composition (%)) (AC), Beta sheet (Chou & Fasman) (BS), Beta turn (Chou & Fasman) (BT), Coil (Deleage & Roux) (C), Hydrophobicity (Aboderin) (H), codon frequencies (CF) [24,25]. Corresponding to each of such graph we have evaluated the fractal dimension using the fractal analysis tools [21]. We have also found the 3D structure of those original and edited sequences. Then we have calculated the fractal dimension of the 3D protein structures given in Table 4 and compared both the FD value through DaliLite Pairwise comparison of protein structures to nd the Z-scores [26] and then find the difference between these two values [27-31].
Table 1. Details of protein Sequence Mutation where in from column we have no. of consecutive current nucleotides and in to column no. of edited nucleotides along with their Position has mentioned.
|
|
Protein Name: OR10K1
|
Protein Name: OR6Y1
|
Protein Name: OR2T4
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
1T
|
1G
|
34
|
4T
|
4G
|
557
|
4G
|
4C
|
841
|
|
|
1C
|
1G
|
7
|
4T
|
4C
|
106
|
4T
|
4A
|
644
|
|
|
3T
|
3A
|
83
|
|
|
|
|
|
|
|
|
Protein Name: OR1C1
|
Protein Name: OR2G2
|
Protein Name: OR2G3
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4T
|
4A
|
766
|
4T
|
4A
|
775
|
4T
|
4G
|
96
|
|
|
3T
|
3A
|
92
|
3C
|
3A
|
884
|
5C
|
5G
|
861
|
|
|
3T
|
3C
|
107
|
2C
|
2A
|
139
|
3T
|
3C
|
833
|
|
|
Protein Name: OR2L2
|
Protein Name: OR2L8
|
Protein Name: OR2T1
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
5T
|
5A
|
147
|
4T
|
4G
|
344
|
4G
|
4C
|
126
|
|
|
5T
|
5C
|
606
|
2T
|
2G
|
9
|
4G
|
4A
|
287
|
|
|
4A
|
4G
|
5
|
2C
|
2G
|
9
|
5C
|
5G
|
485
|
|
|
Protein Name: OR2T3
|
Protein Name: OR2T5
|
Protein Name: OR2T6
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4T
|
4G
|
527
|
5T
|
5A
|
334
|
5T
|
5A
|
539
|
|
|
5C
|
5G
|
503
|
4C
|
4G
|
792
|
4G
|
4A
|
772
|
|
|
4C
|
4T
|
890
|
3C
|
3A
|
155
|
|
|
|
|
|
4C
|
4A
|
653
|
2C
|
2A
|
5
|
|
|
|
|
|
3C
|
3A
|
338
|
|
|
|
|
|
|
|
|
Protein Name: OR5AT1
|
Protein Name: OR5AV1P
|
Protein Name: OR5AX1
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
5C
|
5G
|
851
|
5T
|
5G
|
144
|
5T
|
5G
|
533
|
|
|
5A
|
5T
|
56
|
3T
|
3C
|
60
|
5T
|
5A
|
719
|
|
|
4T
|
4A
|
636
|
3G
|
3C
|
20
|
|
|
|
|
|
3T
|
3C
|
760
|
|
|
|
|
|
|
|
|
3T
|
3A
|
177
|
|
|
|
|
|
|
|
|
3T
|
3G
|
303
|
|
|
|
|
|
|
|
|
2T
|
2G
|
536
|
|
|
|
|
|
|
|
|
Protein Name: OR5AY1
|
Protein Name: OR5BF1
|
Protein Name: OR6F1
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
5T
|
5A
|
504
|
4T
|
4G
|
720
|
5C
|
5G
|
520
|
|
|
4G
|
4A
|
284
|
1C
|
1G
|
88
|
4C
|
4G
|
25
|
|
|
|
|
|
3T
|
3G
|
28
|
3T
|
3G
|
34
|
|
|
Protein Name: OR6K2
|
Protein Name: OR6K3
|
Protein Name: OR6K6
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4T
|
4G
|
759
|
4T
|
4C
|
96
|
4T
|
4G
|
823
|
|
|
4C
|
4G
|
878
|
4A
|
G
|
715
|
4T
|
4A
|
701
|
|
|
3T
|
3G
|
309
|
|
|
|
3T
|
3G
|
156
|
|
|
|
|
|
|
|
|
2C
|
2A
|
15
|
|
|
|
|
|
|
|
|
2G
|
2A
|
901
|
|
|
Protein Name: OR6N1
|
Protein Name: OR6N2
|
Protein Name: OR6P1
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
3T
|
3G
|
427
|
4G
|
4A
|
680
|
4C
|
4T
|
555
|
|
|
4T
|
4A
|
608
|
4T
|
4C
|
726
|
5T
|
5C
|
491
|
|
|
4C
|
G
|
520
|
2T
|
2G
|
28
|
5T
|
5A
|
184
|
|
|
|
|
|
|
|
|
1C
|
1G
|
183
|
|
|
|
|
|
|
|
|
1T
|
1G
|
75
|
|
|
Protein Name: OR10J1
|
Protein Name: OR10J5
|
Protein Name: OR10K2
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4T
|
4G
|
38
|
5T
|
5G
|
629
|
5T
|
5G
|
725
|
|
|
4T
|
4A
|
722
|
4T
|
4A
|
249
|
3T
|
3A
|
280
|
|
|
3C
|
3A
|
78
|
3T
|
3G
|
880
|
|
|
|
|
|
Protein Name: OR10R2
|
Protein Name: OR10T2
|
Protein Name: OR10Z1
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4T
|
4G
|
753
|
5T
|
5C
|
316
|
4T
|
4A
|
497
|
|
|
4A
|
4C
|
187
|
4T
|
4G
|
229
|
4G
|
41
|
709
|
|
|
3T
|
3G
|
677
|
3T
|
3A
|
500
|
|
|
|
|
|
2T
|
2A
|
10
|
2C
|
2A
|
691
|
|
|
|
|
|
Protein Name: OR11L1
|
Protein Name: OR13G1
|
|
|
|
|
Edited
|
From
|
To
|
Position
|
From
|
To
|
Position
|
From
|
To
|
Position
|
|
|
4C
|
4A
|
403
|
4T
|
4C
|
170
|
|
|
|
|
|
3T
|
3G
|
90
|
3T
|
3A
|
634
|
|
|
|
|
|
2C
|
2A
|
626
|
2T
|
2A
|
402
|
|
|
|
Table 2.
|
|
|
(Before) Protein Name: OR10K1
|
|
|
(After)Protein Name: OR10K1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
116
|
28
|
4
|
0
|
1
|
0
|
0
|
|
0
|
116
|
29
|
4
|
0
|
1
|
0
|
0
|
|
0
|
|
T
|
172
|
41
|
13
|
1
|
0
|
0
|
0
|
|
0
|
173
|
40
|
12
|
1
|
0
|
0
|
0
|
|
0
|
|
G
|
98
|
23
|
5
|
1
|
0
|
0
|
0
|
|
0
|
98
|
24
|
5
|
1
|
0
|
0
|
0
|
|
0
|
|
C
|
138
|
54
|
12
|
2
|
0
|
0
|
0
|
|
0
|
138
|
54
|
12
|
2
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 957
|
|
|
|
Total number of Characters : 957
|
|
|
|
|
(Before) Protein Name: OR6Y1
|
|
|
|
(After)Protein Name: OR6Y1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
145
|
25
|
5
|
1
|
0
|
0
|
0
|
|
0
|
145
|
25
|
5
|
1
|
0
|
0
|
0
|
|
0
|
|
T
|
164
|
39
|
10
|
4
|
1
|
1
|
0
|
|
0
|
164
|
39
|
10
|
2
|
1
|
1
|
0
|
|
0
|
|
G
|
107
|
29
|
4
|
2
|
0
|
0
|
0
|
|
0
|
107
|
29
|
4
|
3
|
0
|
0
|
0
|
|
0
|
|
C
|
143
|
56
|
6
|
1
|
0
|
0
|
0
|
|
0
|
143
|
55
|
6
|
1
|
0
|
1
|
0
|
|
0
|
|
Total number of Characters : 994
|
|
|
|
Total number of Characters : 994
|
|
|
|
|
(Before) Protein Name: OR2T4
|
|
|
|
(After)Protein Name: OR2T4
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
142
|
24
|
3
|
0
|
2
|
0
|
0
|
|
0
|
141
|
24
|
3
|
0
|
3
|
0
|
0
|
|
0
|
|
T
|
189
|
26
|
9
|
2
|
3
|
0
|
0
|
|
0
|
189
|
26
|
9
|
1
|
3
|
0
|
0
|
|
0
|
|
G
|
104
|
33
|
9
|
0
|
1
|
0
|
0
|
|
0
|
104
|
33
|
9
|
1
|
1
|
0
|
0
|
|
0
|
|
C
|
129
|
60
|
6
|
4
|
1
|
0
|
0
|
|
0
|
129
|
60
|
6
|
3
|
1
|
0
|
0
|
|
0
|
|
Total number of Characters : 1009
|
|
|
|
Total number of Characters : 1009
|
|
|
|
|
(Before) Protein Name: OR10Z1
|
|
|
(After)Protein Name: OR10Z1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
138
|
14
|
3
|
0
|
0
|
0
|
0
|
|
0
|
136
|
14
|
3
|
0
|
2
|
0
|
0
|
|
0
|
|
T
|
162
|
39
|
8
|
4
|
0
|
0
|
1
|
|
0
|
162
|
39
|
8
|
3
|
0
|
0
|
1
|
|
0
|
|
G
|
106
|
24
|
10
|
3
|
0
|
0
|
1
|
|
0
|
106
|
24
|
10
|
2
|
0
|
0
|
1
|
|
0
|
|
C
|
129
|
53
|
8
|
1
|
1
|
1
|
0
|
|
0
|
129
|
53
|
8
|
1
|
1
|
1
|
0
|
|
0
|
|
Total number of Characters : 957
|
|
|
|
Total number of Characters : 957
|
|
|
|
|
(Before) Protein Name: OR6N2
|
|
|
|
(After)Protein Name: OR6N2
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
129
|
30
|
6
|
1
|
1
|
0
|
0
|
|
0
|
129
|
30
|
6
|
1
|
1
|
0
|
0
|
|
0
|
|
T
|
155
|
45
|
14
|
4
|
1
|
0
|
0
|
|
0
|
155
|
44
|
14
|
4
|
1
|
0
|
0
|
|
0
|
|
G
|
104
|
29
|
4
|
2
|
0
|
0
|
0
|
|
0
|
103
|
29
|
5
|
1
|
0
|
0
|
0
|
|
0
|
|
C
|
135
|
44
|
6
|
1
|
0
|
0
|
0
|
|
0
|
134
|
44
|
6
|
1
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 970
|
|
|
|
Total number of Characters : 970
|
|
|
|
|
(Before) Protein Name: OR5BF1
|
|
|
(After)Protein Name: OR5BF1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
135
|
21
|
4
|
2
|
0
|
0
|
0
|
|
0
|
135
|
21
|
4
|
2
|
0
|
0
|
0
|
|
0
|
|
T
|
165
|
35
|
11
|
4
|
1
|
1
|
0
|
|
1
|
165
|
35
|
10
|
3
|
1
|
1
|
0
|
|
1
|
|
G
|
103
|
32
|
3
|
0
|
1
|
0
|
0
|
|
0
|
104
|
32
|
4
|
1
|
1
|
0
|
0
|
|
0
|
|
C
|
140
|
40
|
9
|
2
|
0
|
0
|
0
|
|
0
|
139
|
40
|
9
|
2
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 954
|
|
|
|
Total number of Characters : 954
|
|
|
|
(Before) Protein Name: OR5AV1P
|
|
|
(After)Protein Name: OR5AV1P
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
128
|
26
|
4
|
1
|
1
|
0
|
0
|
|
0
|
128
|
26
|
4
|
1
|
1
|
0
|
0
|
|
0
|
|
T
|
149
|
40
|
10
|
3
|
5
|
1
|
0
|
|
0
|
149
|
40
|
9
|
3
|
4
|
1
|
0
|
|
0
|
|
G
|
96
|
26
|
6
|
1
|
0
|
0
|
0
|
|
0
|
96
|
26
|
5
|
1
|
0
|
1
|
0
|
|
0
|
|
C
|
110
|
32
|
3
|
3
|
1
|
1
|
0
|
|
0
|
110
|
32
|
5
|
3
|
1
|
1
|
0
|
|
0
|
|
Total number of Characters : 896
|
|
|
|
Total number of Characters : 896
|
|
|
|
|
(Before) Protein Name: OR5AT1
|
|
|
(After)Protein Name: OR5AT1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
124
|
24
|
8
|
1
|
2
|
0
|
0
|
|
0
|
122
|
24
|
8
|
2
|
2
|
0
|
0
|
|
0
|
|
T
|
148
|
48
|
20
|
5
|
0
|
0
|
0
|
|
0
|
146
|
47
|
17
|
4
|
0
|
0
|
1
|
|
0
|
|
G
|
102
|
21
|
3
|
3
|
0
|
0
|
0
|
|
0
|
101
|
22
|
4
|
3
|
0
|
1
|
0
|
|
0
|
|
C
|
133
|
33
|
5
|
1
|
2
|
0
|
0
|
|
0
|
132
|
32
|
5
|
1
|
1
|
1
|
0
|
|
0
|
|
Total number of Characters : 945
|
|
|
|
Total number of Characters : 945
|
|
|
|
|
(Before) Protein Name: OR11L1
|
|
|
(After)Protein Name: OR11L1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
126
|
19
|
6
|
1
|
0
|
0
|
0
|
|
0
|
125
|
20
|
6
|
1
|
1
|
0
|
0
|
|
0
|
|
T
|
164
|
35
|
11
|
3
|
0
|
0
|
1
|
|
0
|
165
|
35
|
11
|
2
|
0
|
0
|
1
|
|
0
|
|
G
|
111
|
33
|
5
|
3
|
0
|
0
|
0
|
|
0
|
111
|
33
|
6
|
3
|
0
|
0
|
0
|
|
0
|
|
C
|
126
|
60
|
8
|
5
|
0
|
0
|
0
|
|
0
|
126
|
59
|
8
|
4
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 985
|
|
|
|
Total number of Characters : 985
|
|
|
|
|
(Before) Protein Name: OR6K6
|
|
|
|
(After)Protein Name: OR6K6
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
132
|
19
|
6
|
0
|
1
|
0
|
0
|
|
0
|
132
|
19
|
6
|
2
|
1
|
0
|
0
|
|
0
|
|
T
|
147
|
39
|
11
|
5
|
2
|
1
|
0
|
|
0
|
147
|
39
|
10
|
3
|
2
|
1
|
0
|
|
0
|
|
G
|
108
|
31
|
6
|
0
|
0
|
0
|
0
|
|
0
|
106
|
30
|
6
|
1
|
1
|
0
|
0
|
|
0
|
|
C
|
115
|
54
|
6
|
2
|
0
|
0
|
0
|
|
0
|
115
|
53
|
6
|
2
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 942
|
|
|
|
Total number of Characters : 942
|
|
|
|
|
(Before) Protein Name: OR2T6
|
|
|
|
(After)Protein Name: OR2T6
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
131
|
19
|
6
|
2
|
0
|
0
|
0
|
|
0
|
130
|
19
|
6
|
3
|
1
|
0
|
0
|
|
0
|
|
T
|
163
|
31
|
7
|
2
|
2
|
0
|
0
|
|
0
|
163
|
31
|
7
|
2
|
1
|
0
|
0
|
|
0
|
|
G
|
98
|
27
|
6
|
7
|
1
|
0
|
0
|
|
0
|
98
|
27
|
6
|
6
|
1
|
0
|
0
|
|
0
|
|
C
|
137
|
40
|
9
|
2
|
2
|
0
|
0
|
|
0
|
137
|
40
|
9
|
2
|
2
|
0
|
0
|
|
0
|
|
Total number of Characters : 942
|
|
|
|
Total number of Characters : 942
|
|
|
|
|
(Before) Protein Name: OR2G2
|
|
|
(After)Protein Name: OR2G2
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
132
|
21
|
7
|
1
|
0
|
0
|
0
|
|
0
|
129
|
20
|
7
|
2
|
2
|
0
|
0
|
|
0
|
|
T
|
161
|
33
|
13
|
5
|
1
|
0
|
0
|
|
0
|
161
|
33
|
13
|
4
|
1
|
0
|
0
|
|
0
|
|
G
|
109
|
30
|
9
|
3
|
0
|
0
|
0
|
|
0
|
109
|
30
|
9
|
3
|
0
|
0
|
0
|
|
0
|
|
C
|
117
|
45
|
14
|
1
|
0
|
0
|
0
|
|
0
|
117
|
44
|
13
|
1
|
0
|
0
|
0
|
|
0
|
|
Total number of Characters : 970
|
|
|
|
Total number of Characters : 970
|
|
|
|
|
(Before) Protein Name: OR2T3
|
|
|
|
(After)Protein Name: OR2T3
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
|
A
|
139
|
17
|
3
|
1
|
0
|
0
|
0
|
|
0
|
137
|
17
|
3
|
3
|
2
|
0
|
1
|
|
0
|
|
T
|
172
|
21
|
7
|
4
|
0
|
1
|
0
|
|
0
|
171
|
21
|
7
|
3
|
0
|
2
|
0
|
|
0
|
|
G
|
126
|
24
|
9
|
0
|
0
|
0
|
0
|
|
0
|
125
|
24
|
9
|
0
|
2
|
0
|
0
|
|
0
|
|
C
|
137
|
54
|
11
|
3
|
4
|
0
|
0
|
|
0
|
137
|
54
|
10
|
3
|
1
|
0
|
0
|
|
0
|
|
Total number of Characters : 973
|
|
|
|
Total number of Characters : 973
|
|
Table 3. Detail no. of nucleotides (A, T, C, and G) consecutively present before and after mutation occurs, along with total no. of nucleotides present in the given amino acid sequences.
|
|
|
(Before) Protein Name: OR6F1
|
|
|
|
(After)Protein Name: OR6F1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
128
|
14
|
7
|
2
|
0
|
0
|
0
|
0
|
128
|
14
|
7
|
2
|
0
|
0
|
0
|
0
|
|
T
|
157
|
31
|
10
|
4
|
1
|
0
|
0
|
0
|
157
|
31
|
10
|
4
|
1
|
0
|
0
|
0
|
|
G
|
113
|
34
|
5
|
1
|
0
|
0
|
0
|
0
|
111
|
33
|
5
|
1
|
1
|
0
|
0
|
1
|
|
C
|
142
|
42
|
7
|
3
|
2
|
0
|
0
|
0
|
142
|
42
|
7
|
2
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 942
|
|
|
Total number of Characters : 942
|
|
|
|
|
(Before) Protein Name: OR10J1
|
|
|
(After)Protein Name: OR10J1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
142
|
27
|
5
|
1
|
0
|
0
|
0
|
0
|
141
|
27
|
5
|
3
|
0
|
0
|
0
|
0
|
|
T
|
152
|
39
|
6
|
4
|
1
|
1
|
0
|
0
|
152
|
39
|
6
|
2
|
1
|
1
|
0
|
0
|
|
G
|
121
|
22
|
7
|
0
|
1
|
0
|
0
|
0
|
120
|
22
|
7
|
0
|
2
|
0
|
0
|
0
|
|
C
|
130
|
38
|
9
|
2
|
1
|
0
|
0
|
0
|
130
|
38
|
8
|
2
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 945
|
|
|
Total number of Characters : 945
|
|
|
|
|
(Before) Protein Name: OR10K2
|
|
|
(After)Protein Name: OR10K2
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
125
|
23
|
4
|
1
|
0
|
0
|
0
|
0
|
124
|
23
|
4
|
2
|
0
|
0
|
0
|
0
|
|
T
|
171
|
45
|
13
|
1
|
2
|
0
|
0
|
0
|
171
|
45
|
12
|
1
|
1
|
0
|
0
|
0
|
|
G
|
100
|
21
|
8
|
1
|
0
|
0
|
0
|
0
|
100
|
21
|
8
|
1
|
1
|
0
|
0
|
0
|
|
C
|
142
|
47
|
7
|
2
|
0
|
0
|
0
|
0
|
142
|
47
|
7
|
2
|
0
|
0
|
0
|
0
|
|
Total number of Characters : 954
|
|
|
Total number of Characters : 954
|
|
|
|
|
(Before) Protein Name: OR6K2
|
|
|
|
(After)Protein Name: OR6K2
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
144
|
20
|
4
|
1
|
0
|
1
|
0
|
0
|
144
|
20
|
4
|
1
|
0
|
1
|
0
|
0
|
|
T
|
156
|
48
|
17
|
4
|
0
|
0
|
1
|
0
|
156
|
48
|
16
|
3
|
0
|
0
|
1
|
0
|
|
G
|
123
|
26
|
3
|
0
|
0
|
0
|
0
|
0
|
123
|
26
|
4
|
2
|
0
|
0
|
0
|
0
|
|
C
|
132
|
43
|
7
|
3
|
1
|
0
|
0
|
0
|
132
|
43
|
7
|
2
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 991
|
|
|
Total number of Characters : 991
|
|
|
|
(Before) Protein Name: OR5AX1
|
|
|
(After)Protein Name: OR5AX1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
130
|
30
|
6
|
2
|
0
|
0
|
0
|
0
|
130
|
30
|
6
|
2
|
1
|
0
|
0
|
0
|
|
T
|
155
|
45
|
15
|
5
|
4
|
0
|
0
|
0
|
155
|
45
|
15
|
5
|
2
|
0
|
0
|
0
|
|
G
|
98
|
30
|
2
|
1
|
0
|
0
|
0
|
0
|
97
|
30
|
2
|
1
|
0
|
1
|
0
|
0
|
|
C
|
118
|
40
|
7
|
1
|
0
|
0
|
0
|
0
|
118
|
40
|
7
|
1
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 960
|
|
|
Total number of Characters : 960
|
|
|
|
|
(Before) Protein Name: OR5AY1
|
|
|
(After)Protein Name: OR5AY1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
127
|
23
|
5
|
1
|
0
|
0
|
0
|
0
|
126
|
22
|
5
|
2
|
0
|
0
|
0
|
1
|
|
T
|
177
|
41
|
10
|
3
|
2
|
0
|
0
|
0
|
177
|
41
|
10
|
3
|
1
|
0
|
0
|
0
|
|
G
|
135
|
21
|
4
|
4
|
0
|
0
|
0
|
0
|
135
|
21
|
4
|
3
|
0
|
0
|
0
|
0
|
|
C
|
138
|
39
|
3
|
1
|
1
|
0
|
0
|
0
|
138
|
39
|
3
|
1
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 960
|
|
|
Total number of Characters : 960
|
|
|
|
|
(Before) Protein Name: OR2T1
|
|
|
|
(After)Protein Name: OR2T1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
139
|
21
|
7
|
0
|
0
|
0
|
0
|
0
|
139
|
21
|
7
|
1
|
0
|
0
|
0
|
0
|
|
T
|
144
|
41
|
13
|
2
|
1
|
0
|
0
|
0
|
144
|
41
|
13
|
2
|
1
|
0
|
0
|
0
|
|
G
|
110
|
27
|
9
|
5
|
0
|
0
|
0
|
0
|
110
|
27
|
9
|
3
|
1
|
0
|
0
|
0
|
|
C
|
118
|
51
|
11
|
0
|
2
|
0
|
0
|
0
|
118
|
51
|
11
|
1
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 973
|
|
|
Total number of Characters : 973
|
|
|
|
|
(Before) Protein Name: OR1C1
|
|
|
|
(After)Protein Name: OR1C1
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
134
|
22
|
2
|
1
|
0
|
1
|
0
|
0
|
133
|
22
|
3
|
1
|
1
|
1
|
0
|
0
|
|
T
|
166
|
37
|
13
|
3
|
1
|
0
|
0
|
0
|
166
|
37
|
11
|
2
|
1
|
0
|
0
|
0
|
|
G
|
122
|
22
|
4
|
2
|
0
|
0
|
0
|
0
|
122
|
22
|
4
|
2
|
0
|
0
|
0
|
0
|
|
C
|
145
|
43
|
4
|
3
|
1
|
1
|
0
|
0
|
145
|
43
|
5
|
3
|
1
|
1
|
0
|
0
|
|
Total number of Characters : 960
|
|
|
Total number of Characters : 960
|
|
|
|
|
(Before) Protein Name: OR2G3
|
|
|
(After)Protein Name: OR2G3
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
122
|
28
|
9
|
1
|
0
|
0
|
0
|
0
|
122
|
28
|
9
|
1
|
0
|
0
|
0
|
0
|
|
T
|
154
|
38
|
12
|
3
|
1
|
1
|
0
|
0
|
154
|
38
|
11
|
2
|
1
|
1
|
0
|
0
|
|
G
|
84
|
37
|
6
|
0
|
0
|
0
|
0
|
0
|
84
|
37
|
6
|
1
|
1
|
0
|
0
|
0
|
|
C
|
138
|
38
|
7
|
2
|
2
|
0
|
0
|
0
|
138
|
38
|
8
|
2
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 945
|
|
|
Total number of Characters : 945
|
|
|
|
|
(Before) Protein Name: OR6K3
|
|
|
|
(After)Protein Name: OR6K3
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
140
|
25
|
4
|
3
|
1
|
1
|
0
|
0
|
140
|
25
|
4
|
2
|
1
|
1
|
0
|
0
|
|
T
|
148
|
45
|
12
|
5
|
1
|
0
|
0
|
0
|
148
|
45
|
12
|
4
|
1
|
0
|
0
|
0
|
|
G
|
104
|
28
|
2
|
1
|
0
|
0
|
0
|
0
|
104
|
27
|
2
|
1
|
0
|
1
|
0
|
0
|
|
C
|
132
|
38
|
11
|
1
|
0
|
1
|
0
|
0
|
130
|
38
|
11
|
1
|
0
|
2
|
0
|
0
|
|
Total number of Characters : 963
|
|
|
Total number of Characters : 963
|
|
|
|
|
(Before) Protein Name: OR2L2
|
|
|
|
(After)Protein Name: OR2L2
|
|
|
occurrences
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
A
|
144
|
25
|
6
|
4
|
0
|
0
|
0
|
0
|
144
|
25
|
6
|
3
|
1
|
0
|
0
|
0
|
|
T
|
158
|
41
|
10
|
2
|
3
|
1
|
0
|
0
|
158
|
41
|
9
|
2
|
1
|
1
|
0
|
0
|
|
G
|
107
|
19
|
5
|
1
|
1
|
0
|
0
|
0
|
107
|
18
|
6
|
1
|
1
|
1
|
0
|
0
|
|
C
|
129
|
38
|
9
|
2
|
0
|
0
|
0
|
0
|
129
|
38
|
9
|
2
|
1
|
0
|
0
|
0
|
|
Total number of Characters : 954
|
|
|
Total number of Characters : 954
|
|
Table 4. Fractal Dimension of 3D Structures Before and After Mutation of nucleotides (A, T, C, G) of the Protein Sequences
|
Protin Name
|
Fractal Value (Before)
|
Fractal Value After
|
Di erence
|
|
OR13G1
|
1.18132
|
1.06183
|
0.11949
|
|
OR10K1
|
1.04287
|
1.09353
|
0.05066
|
|
OR10T2
|
1.10484
|
1.16372
|
0.05888
|
|
OR10R2
|
1.16588
|
1.1053
|
0.06058
|
|
OR6Y1
|
1.07644
|
0.93621
|
0.14023
|
|
OR10K2
|
1.06859
|
1.12591
|
0.05732
|
|
OR2T1
|
1.21768
|
1.22402
|
0.00634
|
|
0R2T4
|
1.03177
|
1.14265
|
0.11088
|
|
OR2T5
|
1.20664
|
1.19132
|
0.01532
|
|
OR2T6
|
1.23737
|
1.20839
|
0.02898
|
|
OR6P1
|
1.11079
|
0.93621
|
0.17458
|
|
OR10Z1
|
1.12791
|
1.00266
|
0.12525
|
|
OR6K2
|
1.03983
|
1.05646
|
0.01663
|
|
OR6K3
|
1.04208
|
1.03747
|
0.00461
|
|
OR6K6
|
1.03636
|
0.93444
|
0.10192
|
|
OR6N1
|
1.10593
|
1.04295
|
0.06298
|
|
OR2L2
|
0.97866
|
1.20928
|
0.23062
|
|
OR6N2
|
1.20545
|
1.18611
|
0.01934
|
|
OR2L8
|
0.96544
|
0.98279
|
0.01735
|
|
OR5BF1
|
1.09345
|
1.06988
|
0.02357
|
|
OR5AX1
|
1.17028
|
1.14738
|
0.0229
|
|
OR5AY1
|
1.23445
|
1.17678
|
0.05767
|
|
OR2G2
|
1.14891
|
1.0137
|
0.13521
|
|
OR6F1
|
1.05202
|
1.07362
|
0.0216
|
|
OR2G3
|
1.06231
|
0.98389
|
0.07842
|
|
OR5AV1
|
1.02043
|
0.96039
|
0.06004
|
|
OR11L1
|
1.18718
|
1.04259
|
0.14459
|
|
OR10J1
|
1.1283
|
1.08652
|
0.04178
|
|
OR2T3
|
1.06912
|
0.98635
|
0.08277
|
|
OR1C1
|
1.0842
|
1.04096
|
0.04324
|
|
OR10J5
|
1.11492
|
1.17187
|
0.05695
|
Results
In this section, we have selected some of OR proteins through which we have studied the effect of local mutation in protein properties as stated above. We have given the accessible residue and alpha helix properties of OR2T6 amino acid sequence before and after mutation in Figures 1 and 2. We have also other amino acid sequences detail has given in Tables 1-3. We have evaluated their fractal dimension of each of its properties. We have given the 3D structure of OR2T6 sequence after and before mutation in Figure 3, also marked the changes in 3D structure of before and after mutation. We have also generated 3D structure of each of the amino acid sequence, and also evaluate the fractional dimension of each of the structures before and after mutation.
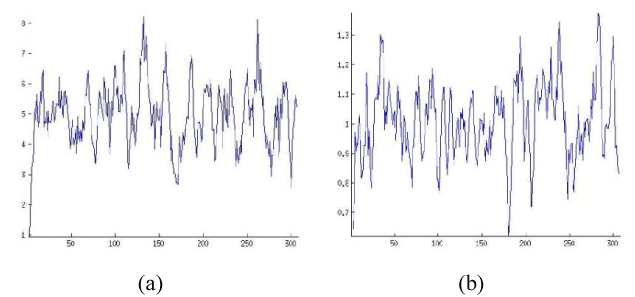
Figure 1. Protein Properties of OR2T6 protein sequence ORs
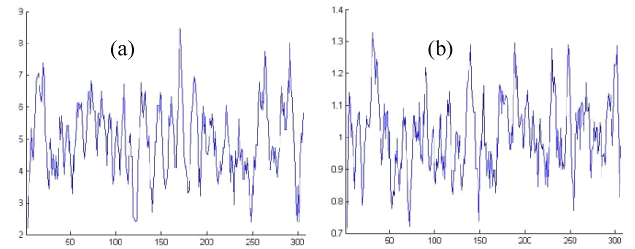
Figure 2. Edited Protein Properties of OR2T6 protein sequence ORs
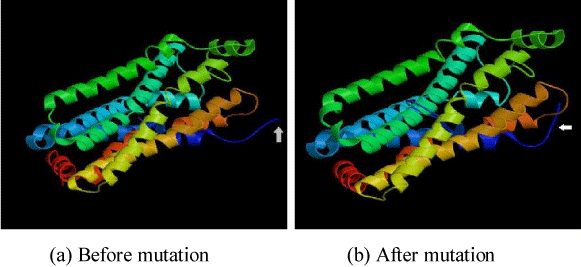
Figure 3. Change in structure of OR2T6 protein sequence before and after mutation
In the previous section a set of 32 OR sequences has been experimented through Fractal dimensional analysis. Mainly we are intended to observe the similarities or dissimilarities in the original and edited (mutated) OR sequences. We have found out the fractal dimension of the different graphs and their corresponding fractal dimension of the protein properties as mentioned earlier.
It is obvious that if the percentage of Accessible Residues (AR) is less than the percentage of Buried Residues (BR) must be higher as these two protein properties are complementary to each other. Accordingly, the other protein properties also get affected due to the decrement of percentage of AR in an OR protein. In other words, all such protein properties are interlinked for a protein. This particular detail has been wide-opened in the Table 1.
Here we have considered 13 OR sequences out of those 31 sequences for which a detail analysis has been made in the Tables 5 and 6.
Table 5. AR - Accessible residue BR- Buried residue Properties AA - Amino Acid AH - Alpha Helix properties
|
|
|
AR
|
|
|
BR
|
|
|
AH
|
|
|
AA
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
protein
|
A
|
B
|
R
|
A
|
B
|
R
|
A
|
B
|
R
|
A
|
B
|
R
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
OR10K1
|
1.94359
|
1.94361
|
B=A
|
1.94266
|
1.94331
|
B<A
|
1.9427
|
1.94318
|
B<A
|
1.94289
|
1.94358
|
B<A
|
|
OR10Z1
|
1.94373
|
1.94284
|
B>A
|
1.94281
|
1.9432
|
B<A
|
1.94253
|
1.94298
|
B<A
|
1.94331
|
1.94303
|
B>A
|
|
OR2G3
|
1.94264
|
1.94372
|
B<A
|
1.94283
|
1.94315
|
B<A
|
1.94266
|
1.94299
|
B<A
|
1.94247
|
1.94366
|
B<A
|
|
OR2T6
|
1.94346
|
1.9431 B>A
|
1.94263
|
1.94317
|
B<A
|
1.94314
|
1.94269
|
B<A
|
1.94324
|
1.94299
|
B>A
|
|
OR2T1
|
1.94293
|
1.9433 B<A
|
1.9426
|
1.94308
|
B>A
|
1.94179
|
1.94269
|
B<A
|
1.94325
|
1.9431
|
B<A
|
|
OR6N2
|
1.94274
|
1.9436 B<A
|
1.94265
|
1.94324
|
B<A
|
1.94367
|
1.94326
|
B>A
|
1.94284
|
1.94389
|
B<A
|
|
OR6P1
|
1.9431
|
1.94313
|
B=A
|
1.94232
|
1.94333
|
B<A
|
1.94303
|
1.94297
|
B=A
|
1.94215
|
1.94378
|
B<A
|
|
OR13G1
|
1.94248
|
1.94283
|
B<A
|
1.94251
|
1.9434 B<A
|
1.94319
|
1.94306
|
B<A
|
1.94325
|
1.94259
|
B>A
|
|
OR6K2
|
1.94279
|
1.94315
|
B<A
|
1.9424
|
1.94309
|
B<A
|
1.9431
|
1.94336
|
B<A
|
1.94282
|
1.94355
|
B<A
|
|
OR2T5
|
1.94302
|
1.94346
|
B<A
|
1.94248
|
1.94357
|
B<A
|
1.9427
|
1.9432
|
B<A
|
1.94292
|
1.94342
|
B<A
|
|
OR10J5
|
1.94305
|
1.94333
|
B<A
|
1.94182
|
1.94332
|
B<A
|
1.94294
|
1.94341
|
B<A
|
1.94331
|
1.9435
|
B<A
|
|
OR6Y1
|
1.94263
|
1.94321
|
B<A
|
1.94215
|
1.94283
|
B<A
|
1.94279
|
1.94317
|
B<A
|
1.94237
|
1.94334
|
B<A
|
|
OR1C1
|
1.94271
|
1.94321
|
B<A
|
1.94173
|
1.94304
|
B<A
|
1.94244
|
1.94273
|
B<A
|
1.94225
|
1.94333
|
B<A
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 6. BS- Beta sheet BT-Beta turn HPC - Hydrophobicity coil, codon properties, and A, B , R used for After , Before and Result respectively
|
11
|
|
|
BS
|
|
|
BT
|
|
|
Coil
|
|
|
HPC
|
|
|
codons
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
protein
|
A
|
B
|
R
|
A
|
B
|
R
|
A
|
B
|
R
|
A
|
B
|
R
|
A
|
B
|
R
|
|
|
|
OR10K1
|
1.94326
|
1.94386
|
B<A
|
1.94292
|
1.94295
|
B=A
|
1.9447
|
1.94411
|
A>B
|
1.9428
|
1.94275
|
B<A
|
1.94279
|
1.94336
|
B<A
|
|
|
|
OR10Z1
|
1.94289
|
1.94243
|
B>A
|
1.94337
|
1.94271
|
B>A
|
1.94379
|
1.94409
|
B<A
|
1.94266
|
1.94276
|
B<A
|
1.94319
|
1.942298
|
B>A
|
|
|
|
OR2G3
|
1.94239
|
1.94389
|
B<A
|
1.94295
|
1.94247
|
B>A
|
1.94407
|
1.94395
|
B>A
|
1.94263
|
1.94239
|
B>A
|
1.94278
|
1.94331
|
B<A
|
|
|
|
OR2T6
|
1.94233
|
1.9439
|
B<A
|
1.94289
|
1.94252
|
B>A
|
1.94407
|
1.94398
|
B=A
|
1.94289
|
1.94305
|
B<A
|
1.94307
|
1.94279
|
B>A
|
|
|
|
OR2T1
|
1.94279
|
1.94307
|
B<A
|
1.94216
|
1.94254
|
B<A
|
1.94374
|
1.94401
|
B<A
|
1.94267
|
1.94299
|
B<A
|
1.94319
|
1.94302
|
B>A
|
|
|
|
OR6N2
|
1.94281
|
1.94385
|
B<A
|
1.94367
|
1.94291
|
B>A
|
1.94423
|
1.94408
|
B>A
|
1.943
|
1.94276
|
B>A
|
1.94238
|
1.94295
|
B<A
|
|
|
|
OR6P1
|
1.94243
|
1.94387
|
B<A
|
1.94305
|
1.94279
|
B>A
|
1.94407
|
1.94414
|
B<A
|
1.944266
|
1.94276
|
B<A
|
1.94231
|
1.94362
|
B<A
|
|
|
|
OR13G1
|
1.94236
|
1.94273
|
B<A
|
1.94336
|
1.94327
|
B>A
|
1.94409
|
1.94444
|
B<A
|
1.9425
|
1.94307
|
B>A
|
1.94359
|
1.94289
|
B>A
|
|
|
|
OR6K2
|
1.94289
|
1.94243
|
B>A
|
1.9427
|
1.94261
|
B>A
|
1.94379
|
1.94412
|
B<A
|
1.94253
|
1.94278
|
B<A
|
1.94284
|
1.94331
|
B<A
|
|
|
|
OR2T5
|
1.94287
|
1.9432
|
B<A
|
1.94303
|
1.94268
|
B>A
|
1.94407
|
1.94405
|
B=A
|
1.9428
|
1.94316
|
B<A
|
1.94242
|
1.94321
|
B<A
|
|
|
|
OR10J5
|
1.94231
|
1.94381
|
B<A
|
1.94289
|
1.9428
|
B=A
|
1.94417
|
1.944
|
B>A
|
1.94187
|
1.94323
|
B<A
|
1.94315
|
1.94352
|
B<A
|
|
|
|
OR6Y1
|
1.94273
|
1.94373
|
B<A
|
1.9426
|
1.94367
|
B>A
|
1.944
|
1.94389
|
B>A
|
1.94334
|
1.94281
|
B>A
|
1.94263
|
1.94281
|
B<A
|
|
|
|
OR1C1
|
1.94246
|
1.94389
|
B<A
|
1.94296
|
1.94256
|
B>A
|
1.94413
|
1.94408
|
B=A
|
1.94211
|
1.94259
|
B<A
|
1.94179
|
1.94361
|
B<A
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Let us illustrate the above fact by taking one example.
In case of OR2T6, the mutated sequence differs from the original OR2T6 in two positions at 539 and 772 of the sequence. The poly-string of 5Ts has been changed into 5As in the position 539 in the edited sequence and in position 772, 4Gs has been changed into 4As. Consequently the frequency of 1As is decreases by 1, 4As increased by 1, 5As increased by 1, 5Ts decreased by 1 and 4Gs decreased by 1 ,but the other frequencies of poly-strings has been remain unchanged even though the above changes in the poly-string frequencies of 5Ts and 4Gs.
As a result, the Fra2021 Copyright OAT. All rights reserv Residues (AR) in the mutated sequences is less than that of the original OR2T6. As a result, the FD of Buried Residues (BR) for the mutated OR2T6 is greater than that of the original, the FD of Alpha helix is greater than the and even after mutation the FD coil are almost same as it was in original OR2T6 whereas the FD of Amino Acid Composition (AA), Codon frequency (C) and Beta Turn (BT) of the mutated OR2T6 is less than that of the OR2T6. The FD of Beta Sheet and Hydrophobicity (HPC) is greater than that of the original OR2T6. Also the tertiary structures of the protein for the OR2T6 and corresponding edited sequence have been changed. The fractal dimension of those tertiary structure of proteins are 1:23737 and 1:20839 respectively.
In this study, we have considered a few OR sequences to understand the local mutation in the protein level. The detail account of poly-string frequencies have been made for each of the ORs and corresponding edited sequences. We then saw the effect of those local changes in poly-string frequency through the protein properties and tertiary structure of the protein of each of the ORs and edited sequences. As we illustrated through the example of OR2T 6 and its edited sequence in the discussion, we found that the edited sequence is almost same with the original OR2T 6 in the sense of ordering of the nucleotide bases except for the frequency change in case of 1As, 4As, 5As, 5Ts and 4Gs. Here we have found that the edited sequence of OR2T 6 is almost sequentially similar and also the tertiary structure is almost similar to the original tertiary structure of the protein OR2T6. Hence, it is our strong conviction that these kinds of mutation can be allowed in making the function of OR2T6 un-altered. Of course, the biological experiment is left to make us assured about its original functionality of OR2T6. Similar kind of analysis is also applicable for the other sequences in accordance with the change and their effect in protein properties and their tertiary structures.
- Fiser A (2004) Protein structure modeling in the proteomics era. Expert Rev Proteomics 1: 97-110. [Crossref]
- Gaillard I, Rouquier S, Giorgi D (2004) Olfactory receptors. Cell Mol Life Sci 61: 456-469. [Crossref]
- DNA to protein translation, http://insilico.ehu.es/translate/
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162-1164. [Crossref]
- Dunbrack RL Jr, Karplus M (1994) Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat Struct Biol 1: 334-340. [Crossref]
- Vásquez M (1996) Modeling side-chain conformation. Curr Opin Struct Biol 6: 217-221. [Crossref]
- Wagner GC, Colvin JT, Allen JP, Stapleton HJ (1985) Fractal models of protein structure, dynamics and magnetic relaxation. Journal of the American Chemical Society 107: 5589-5594.
- Delarue M, Koehl P (1997) The inverse protein folding problem: self-consistent mean field optimization of a structure specific mutation matrix. Pac Symp Biocomput. [Crossref]
- Levitt M, Gerstein M, Huang E, Subbiah S, Tsai J (1997) Protein folding: the endgame. Annu Rev Biochem 66: 549-579. [Crossref]
- Xiang Z, Honig B (2001) Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol 311: 421-430. [Crossref]
- Malnic B, Godfrey PA, Buck LB (2004) The human olfactory receptor gene family. Proc Natl Acad Sci U S A 101: 2584-2589. [Crossref]
- Wu G, Fiser A, ter Kuile B, Sali A, Müller M (1999) Convergent evolution of Trichomonas vaginalis lactate dehydrogenase from malate dehydrogenase. Proc Natl Acad Sci U S A 96: 6285-6290. [Crossref]
- Chothia C, Lesk AM (1986) The relation between the divergence of sequence and structure in proteins. EMBO J 5: 823-826. [Crossref]
- Schmitt C, Sanchez C, Desobry-Banon S, Hardy J (1998) Structure and technofunctional properties of protein-polysaccharide complexes: a review. Crit Rev Food Sci Nutr 38: 689-753. [Crossref]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275-282. [Crossref]
- Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33: W306-310. [Crossref]
- Boyer RS, Moore JS (1977) "A Fast String Searching Algorithm.” Comm. ACM (New York, NY, USA: Association for Computing Machinery) 20: 762772.
- Baeza-Yates R, Gonnet GH (1992) A new approach to text searching, Com-munications of the ACM, 35: 74-82.
- Karp RM, Rabin MO (1987) E cient randomized pattern-matching algo-rithms. IBM J Res Dev 31: 249-260.
- Ponder JW, Richards FM (1987) Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol 193: 775-791. [Crossref]
- Cohen I, Herlin I (1998) Curves matching using geodesic paths. In CVPR98. IEEE, Santa-Barbara, USA, pp. 741746.
- Stein JM (1975) The effect of adrenaline and of alpha- and beta-adrenergic blocking agents on ATP concentration and on incorporation of 32Pi into ATP in rat fat cells. Biochem Pharmacol 24: 1659-1662. [Crossref]
- Conversion of protein structure from Amino acid sequences http://ps2.life.nctu.edu.tw/index.php/
- Janin J, Wodak S (1978) Conformation of amino acid side-chains in proteins. J Mol Biol 125: 357-386. [Crossref]
- Feyfant E, Sali A, Fiser A (2007) Modeling mutations in protein structures. Protein Sci 16: 2030-2041. [Crossref]
- Pairwise alignment of protein structures http://www.ebi.ac.uk/Tools/dalilite/
- Holm L, Park J (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16: 566-567. [Crossref]
- Malnic B, Godfrey PA, Buck LB (2004) The human olfactory receptor gene family. Proc Natl Acad Sci U S A 101: 2584-2589. [Crossref]
- Benoit B (1975) Mandelbrot. Les objects fractals: forme, hassard et dimesion, Flammarion, Paris.
- Hassan SS, Choudhury PP, Guha R, Chakraborty S, Goswami A (2010) "DNA Sequence Evolution through Intregral Value Transformations". Interdiscip Sci 4: 128-132. [Crossref]



