Abstract
In all aerobic organisms on Earth, molecular oxygen is essential for cellular respiration, and our body consisting of those cells is continuously exposed to “oxidative stress”. In response to oxidative stress, NF-E2-related transcription factor (Nrf2) encoded by the Nrf2 gene is considered to play a pivotal role in cellular defense via transcriptional up-regulation of many downstream genes, including those for metabolizing enzymes and transporters essential for cellular function. However, it is also true that the Nrf2 gene is regarded as a double-edged sword. While it plays a role in protecting normal cells from external toxic challenges and oxidative stress, the Nrf2 gene can also endow cancer cells resistance to anticancer drugs. The Nrf2 protein interacts with the antioxidant responsive element (ARE) located in the promoter region of the Nrf2 gene as well as its downstream target genes. Genetic polymorphisms and/or mutations in the Nrf2 gene have hitherto been identified in human samples, one single nucleotide polymorphism (SNP; -617C>A; rs6721961) in the ARE-like loci of the human Nrf2 gene reportedly affects the positive feedback loop of transcriptional activation of the Nrf2 gene. Ethnic group-dependent difference is observed for that SNP, where the frequency of the -617A allele is high in Japanese, Taiwanese, and Chinese populations. These allele frequency differences in the Nrf2 gene may reflect genetic alterations and selection that took place during inter-continental migrations of Homo sapiens.
Key words
Nrf2, lung, cancer, chronic obstructive pulmonary disease (COPD), oxidative stress, reactive oxygen species (ROS), heme oxygenase-1 (HO-1)
Introduction
The history of the universe including time, space, and matters began shortly after the “Big Bang”, 13.7 billion years ago. About 9 billion years later, our solar system was formed in the Milky Way Galaxy. Oxygen was essentially absent from the atmosphere of Earth when this planet was first created about 4.54 billion years ago [1]. Carbon dioxide (CO2) was the major gas present in the atmosphere of early Earth. After the chemical evolution from the RNA-protein world to the DNA/RNA-protein world, anaerobic microbes were created as original life at least from 3.8 billion to 4.1 billion years ago [2-4]. It is speculated that molecular oxygen (O2) was first produced somewhere in oceans of Earth around 2.7 billion to 2.8 billion years ago, and then it emerged in atmosphere around 2.45 billion years ago (Figure 1). The origin of molecular oxygen in Earth's atmosphere derived from the biological activity of tiny organisms, namely cyanobacteria that appeared on earth about 3.5 billion years ago [5]. Cyanobacteria were capable to produce carbohydrates from water and carbon dioxide molecules by using the photon energy of sunshine. During the process of photosynthesis, molecular oxygen was produced as exhaust gas. Cyanobacteria are considered to have given rise to plant chloroplasts. As the plants world prospered on Earth and photosynthesis became more common, the rate of O2 production increased until the present concentration of oxygen in the atmosphere reached about 580 million years ago.
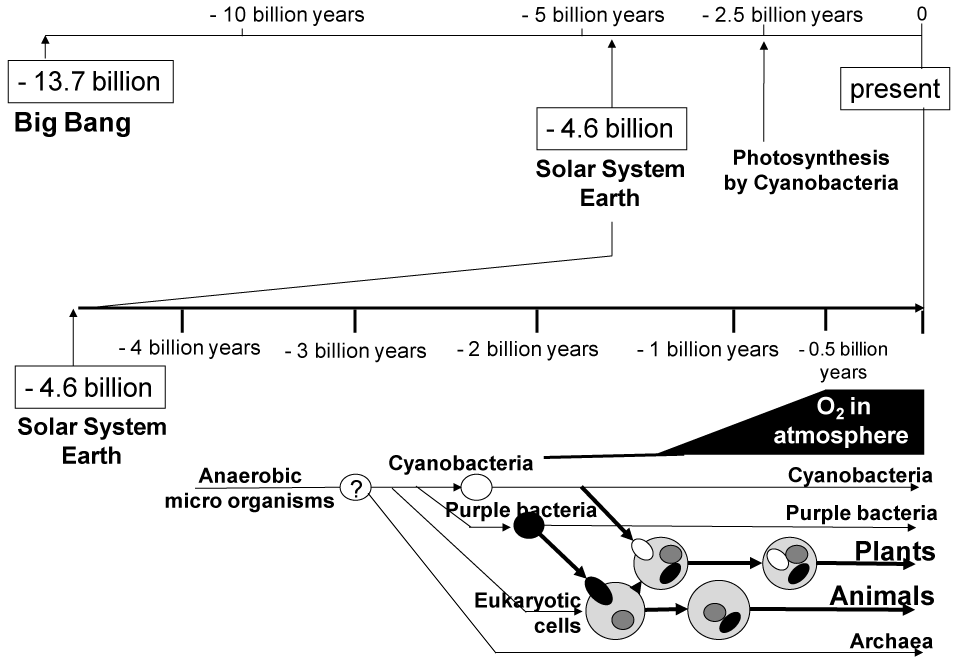
Figure 1. The evolution of organisms on Earth and the emergence of molecular oxygen (O2) in the history of the universe after Big Bang.
Today, molecular oxygen is essential for cellular respiration in all aerobic organisms, where mitochondria generate ATP via oxidative phosphorylation. In contrast, to anaerobic organisms, molecular oxygen is toxic and harmful. It was one of the evolutionally critical points that anaerobes incorporated purple bacteria to acquire high efficiencies of ATP production via oxidative phosphorylation. Purple bacteria are considered to have introduced the ancestor responsible for eukaryotic mitochondria. Mitochondria have their own DNA to proliferate in host eukaryotic cells. Since mitochondria-hosting eukaryotic cells (i.e., aerobic cells) had originated anaerobic microbes, those cells started to encounter the risk of oxygen toxicity. Indeed, reactive oxygen (ROS) species generated from molecular oxygen are causal factors in the production of cell damage as well as the mutation of genomic DNA. ROS which consist of superoxide anion radical (.O2-), hydroxyl radical (.OH) and hydrogen peroxide (H2O2) play important roles in many essential cellular physiological functions, including growth, differentiation, apoptosis, and aging [6]. Under normal physiological conditions, there is a balance between oxidants and antioxidants, or reduction-oxidation (redox) homeostasis.
One of the classical but important mechanisms counteracting ROS is the glutathione (GSH) system [7,8]. GSH is a ubiquitous tripeptide thiol (L--glutamyl-L-cysteinyl-glycine). It is a vital intra- and extra-cellular protectant [9,10] and an effective scavenger of ROS. It is estimated that GSH biosynthesis originated about 3 billion years ago. GSH is found in the vast majority of eukaryotes, whereas in eubacteria, GSH biosynthesis is limited to only two groups, namely cyanobacteria and purple bacteria that are thought to be the ancestors of chloroplast and mitochondria, respectively [11].
Oxidative stress in pathophysiology
Our body consisting of eukaryotic cells is continuously exposed to “oxidative stress”. The term “oxidative stress” is defined as a disturbed balance between the production of ROS and the ability of the body to counteract or detoxify their harmful effects through neutralization by antioxidants and enzymatic reactions [12]. This biochemical phenomenon is inevitable as long as we live on Earth. Oxidative stress leads to many pathophysiological conditions in our body. Some of these include cancers, chronic obstructive pulmonary disease (COPD), atherosclerosis, hypertension, diabetes mellitus, and neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease.
The lung is the organ that is continuously exposed to atmosphere’s molecular oxygen and has a risk of oxidative damage. Indeed, accumulating evidence strongly suggests that oxidative stress and oxidative damage are involved in the pathogenesis of COPD, where oxidative stress is considered to cause direct injury and inflammation in the respiratory tract, lung, and other organs and cell types. The sources of the increased oxidative stress in the respiratory compartment of COPD patients derive from the increased burden of oxidants from environmental exposures, such as cigarette smoke. Furthermore, ROS and reactive nitric oxide (NO) are also released from leukocytes and macrophages involved in the inflammatory process in the lungs of COPD patients. ROS can induce lipid peroxidation and yield products such as 4-hydroxynonenal and malondialdehyde to stimulate pulmonary inflammation. Increased levels of lipid peroxidation products were detected in the breath condensate and plasma of smokers and patients with stable COPD [13]. Antioxidant defenses, including GSH, vitamin E, glutathione peroxidase, superoxide dismutase, and heme oxygenase-1 (HO-1), are of importance to reduce oxidative damage in the lung.
Key role of heme oxygenase-1 to suppress pulmonary inflammation
HO-1 is a rate-limiting enzyme in the metabolic pathway of heme catabolism to generate biliverdin that is successively metabolized to bilirubin by the enzymatic action of biliverdin reductase. Bilirubin, thus formed, exhibits anti-oxidative, anti-apoptotic, and anti-inflammatory activities. Decrease in the HO-1 level is reportedly associated with the pathogenesis of some age-dependent disorders, including COPD and cancer. It is hypothesized that enhanced expression of HO-1 provides an anti-inflammatory effect and confers cytoprotection. To examine this hypothesis, Shinohara et al. [14] examined whether HO-1 overexpression would prevent pulmonary emphysemia induced by porcine pancreatic elastase (PPE). They inoculated mice with an adenovirus encoding HO-1 in the lung [14]. In fact, adenovirus-mediated overexpression of HO-1 in the lung significantly attenuated PPE-induced increase of neutrophils in bronchoalveolar lavage fluid and enlargement of alveoli. In addition, HO-1 overexpression reduced PPE-induced TNFα, IL-6, and keratinocyte-derived chemokine in bronchoalveolar lavage fluid. Furthermore, Sato et al. [15] have confirmed the anti-inflammatory effect of HO-1 using a mouse silicosis model where HO-1 expression was up-regulated by treatment with hemin [15]. Hemin is a strong inducer for HO-1 gene expression, and NF-E2-related transcription factor (Nrf2) induces HO-1 gene expression, as described below. Other relevant studies have also proven that HO-1 is a key player in the suppression of pulmonary inflammation. HO-1 inhibits IL-13-induced goblet cell hyperplasia associated with CLCA1 suppression in normal human bronchial epithelial cells, as well [16].
Nrf2-mediated transcriptional regulation
Nrf2 is a master switch for transcriptional up-regulation of a variety of target genes, including those for metabolizing enzymes (e.g., HO-1) and transporters essential for cellular defense in response to oxidative and/or electrophilic stress [17-19]. The Nrf2 protein is a basic region-leucine zipper (bZip)-type transcription factor, and it targets the anti-oxidant response element (ARE) with the consensus sequence of 5’- A/GTGACNNNGC) [19,20] This Nrf2 system has been evolutionarily conserved among different species [21].
The activation and nuclear translocation processes of Nrf2 seem to be complex. As shown in Figure 2, at least three distinct pathways may be involved in the activation of the Nrf2 protein leading to HO-1 induction: oxidation of critical cysteinyl residues of the Keap1 protein and concomitant inhibition of ubiquitination activity of Keap1 (Pathway A); phosphorylation of the Nrf2 protein via protein kinases, such as p38MAPK, PI3K, PKC, and PERK (Pathway B); direct binding of heme to Bach1 and facilitation of Nrf2/small Maf heterodimer formation (Pathway C).
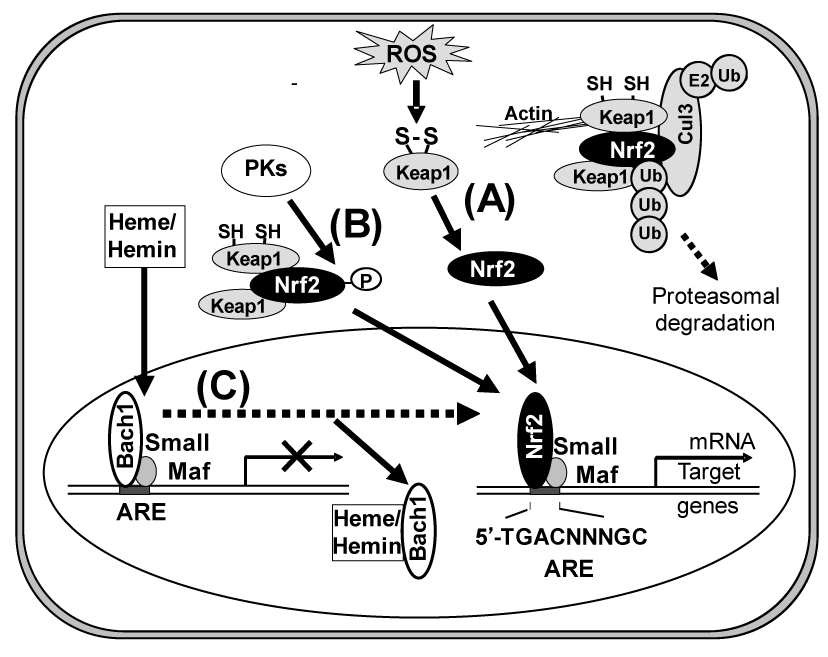
Figure 2. Activation of Nrf2 via three different pathways (A, B, and C). Pathway A, under homeostatic conditions, Nrf2 is sequestered in the cytoplasm by the Keap1-Cul3 complex and rapidly degraded in a ubiquitin-proteasome-dependent manner. After an oxidative challenge, oxidation of two reactive cysteine residues of Keap1 inhibits the ubiquitination reaction of Nrf2 mediated by the Keap1-Cul3 complex, which results in both cytoplasmic accumulation and nuclear translocation of Nrf2. Pathway B, activation of Nrf2 is mediated by protein kinases (PKs), such as p38MAPK, PI3K, PERK, and PKC. Pathway C, under normal conditions, the chromatin structure of many target genes is in a pre-activation state, but transcription is repressed by Bach1. Heme or hemin binds to Bach1, inhibiting its DNA binding activity and inducing its nuclear export. In the nuclei, the activated Nrf2 dimerizes with small Maf nuclear protein (MafK) for effective binding to the ARE consensus sequence in the promoter region of Nrf2-target genes, such as HO-1 and g-glutamyl cysteine ligase.
The HO-1 induction is dependent on transcription and de novo protein synthesis, and it is preceded by the nuclear accumulation of the Nrf2 transcription factor. In pathway A, Nrf2 is repressed under quiescent conditions, where Keap1 and Cul3 constitute a unique ubiquitin E3 ligase that leads to the degradation of Nrf2. Upon exposure to oxidants/electrophiles, the enzymatic activity of this ligase complex is inhibited and the complex fails to degrade Nrf2, resulting in the transcriptional activation of Nrf2 target genes [22,23]. Cys151 of Keap1 reportedly plays an important role to facilitate Nrf2 activation in response to oxidants/electrophiles [23].
Regarding pathway B, Martin et al. [24] reported that HO-1 expression is regulated through the PI3K/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. Kocanova et al. [25] more recently characterized the signaling pathways and the mechanisms leading to the up-regulation of HO-1 in cancer cells subjected to hypericin-based photodynamic therapy. They have shown that HO-1 induction mechanisms involve the p38MAPK and PI3K signaling cascade. Besides p38MAPK and PI3K, the activation of Nrf2 is mediated by other protein kinases, such as PERK and PKC, being dependent on cell types [26-31].
In pathway C, heme or hemin regulates the dynamic exchange of Bach1 and Nrf2 in the Maf transcription factor network. Igarashi and his colleagues proposed this direct interaction model [32-35]. The transcription repressor Bach1 is a sensor and effector of heme/hemin that regulates the expression of HO-1 and globin genes [36-38]. Under normal conditions, the chromatin structure of HO-1 is in a preactivation state, but transcription is repressed by Bach1. Heme/hemin binds to Bach1, inhibiting its DNA binding activity [39] and inducing its nuclear export [40]. Furthermore, heme/hemin induce ubiquitination and degradation of the transcription factor Bach1 [41]. As a consequence, heme/hemin induces the switching of Nrf2/small Maf hetero-dimers, resulting in HO-1 expression [42].
SNP (-617C>A; rs6721961) in the Nrf2 gene
Yamamoto et al. first reported the structure of the Nrf2 gene and found three SNPs (-653A>G, -651G>A, and -617C>A) and one triplet repeat polymorphism in its regulatory region [43] (Figure 3). Three years later, Marzec et al. examined the impact of those SNPs on the regulation of Nrf2 gene expression [44]. In transient transfection assays, they found that the -617C>A SNP significantly affects basal Nrf2 protein levels and its function in vitro [44]. SNP -617C>A was found to be associated with a higher risk of oxidant-induced acute lung injury in humans [44]. It is likely that the SNP (c.-617C>A) in the ARE-like loci of the human Nrf2 gene is important for self-induction of the Nrf2 gene (Figure 4). To our knowledge, the report of Marzec et al. [44] is the first one demonstrating that functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury.
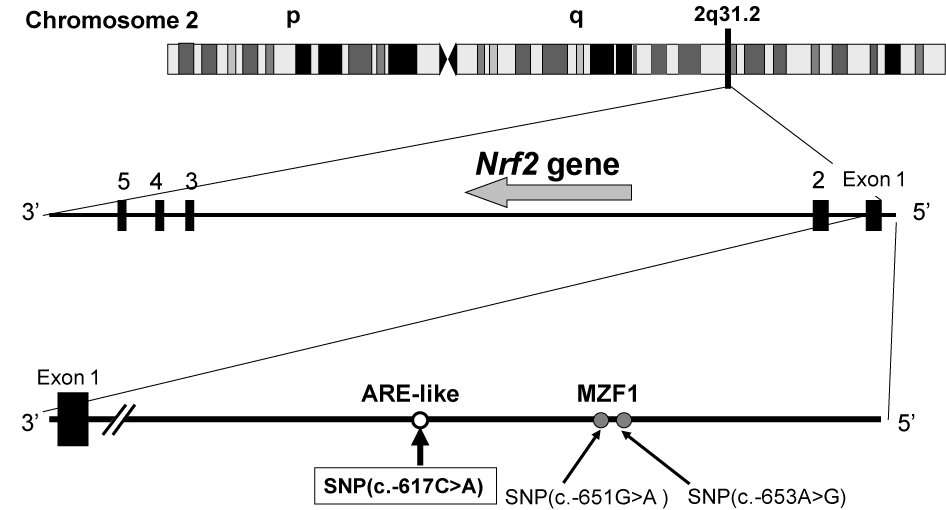
Figure 3. SNP (- 617C>A) in the promoter region of the human Nrf2 gene located on chromosome 2q31.2.
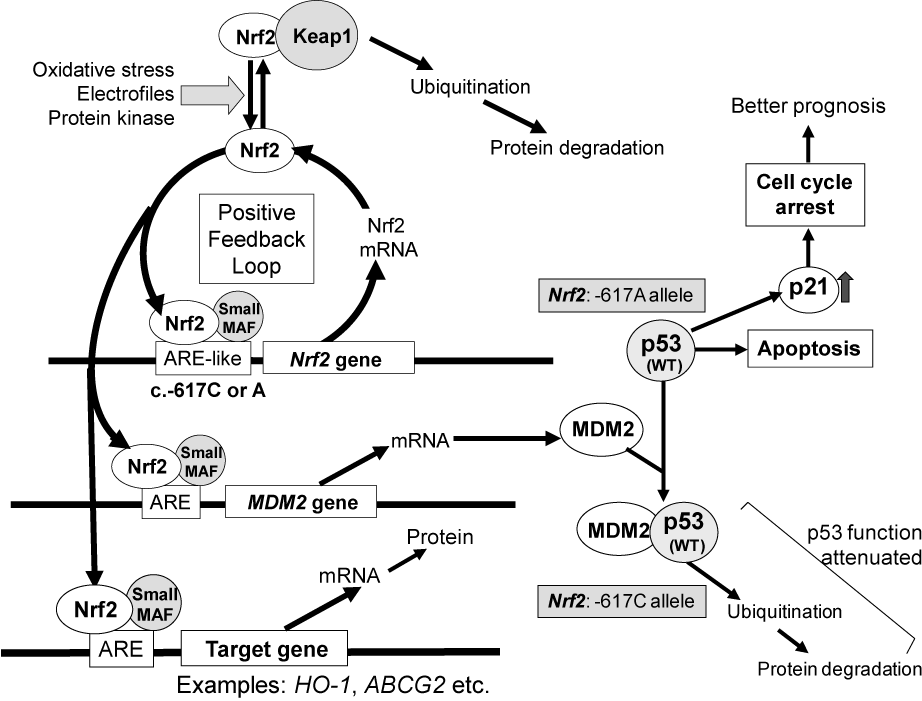
Figure 4. Schematic illustrations showing the effect of NRF2 SNP -617C>A and MDM2 gene on the p53-mediated suppression of cancer cell proliferation and ABCG2-mediated drug resistance to gefitinib [50].
Interestingly, ethnic group-dependent difference was observed in the Nrf2 genotype, where the frequency of the -617A allele is high in Japanese, Taiwanese, and Chinese populations (Table 1). The ethnic group-dependent difference may reflect genetic alterations and selection that took place in the Nrf2 gene during inter-continental migrations of Homo sapiens (Figure 5). With this respect, the authors have recently reported that inter-continental migration of Homo sapiens is well associated with ethnic-dependent differences in one SNP 538G>A (rs17822931: Gly180Arg) in the human ABCG11 gene [45].
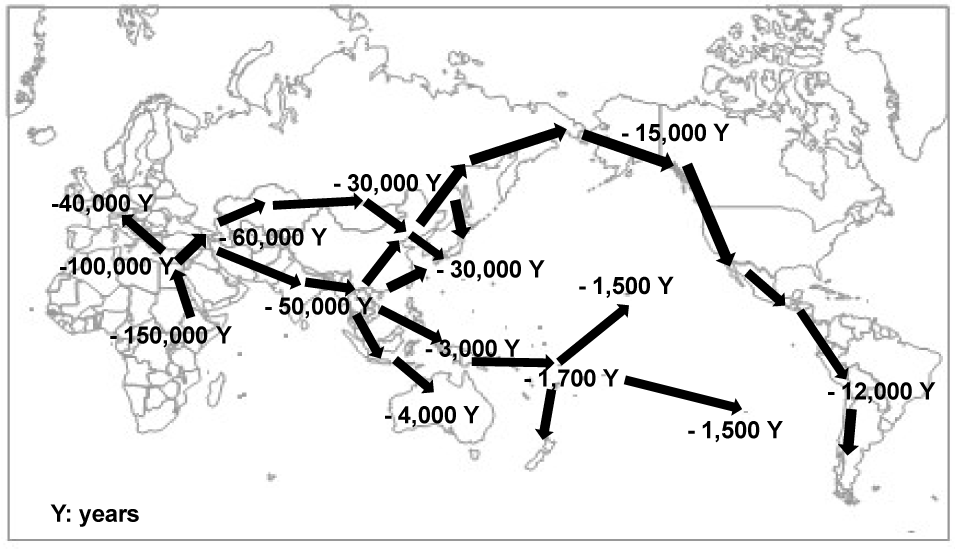
Figure 5. Schematic illustration of inter-continental migration of Homo sapiens [45].
Table 1. Frequencies of wild type (-617C) and SNP (-617A) alleles in the Nrf2 gene among different ethnic groups
|
Allele frequency
|
Nrf2 (-617)
|
|
Ethnic group |
C |
A |
C/C |
C/A |
A/A |
N |
Data source |
African |
0.925 |
0.075 |
0.850 |
0.150 |
0.000 |
246 |
[64] |
African-American |
0.893 |
0.107 |
0.787 |
0.213 |
0.000 |
61 |
[64] |
European |
0.883 |
0.117 |
0.778 |
0.208 |
0.013 |
379 |
[64] |
American in Utah |
0.888 |
0.112 |
0.788 |
0.200 |
0.012 |
85 |
[64] |
American mixed |
0.862 |
0.138 |
0.757 |
0.210 |
0.033 |
181 |
[64] |
Mexican in Los Angels |
0.803 |
0.197 |
0.667 |
0.273 |
0.061 |
66 |
[64] |
Japanese |
0.775 |
0.225 |
0.618 |
0.315 |
0.067 |
89 |
[64] |
Japanese (lung cancer) |
0.748 |
0.252 |
0.558 |
0.380 |
0.062 |
387 |
[50] |
Taiwanese |
0.726 |
0.274 |
0.524 |
0.405 |
0.071 |
168 |
[50] |
Chinese in Beijing |
0.722 |
0.278 |
0.515 |
0.412 |
0.072 |
97 |
[64] |
Southern Han Chinese |
0.710 |
0.290 |
0.500 |
0.420 |
0.080 |
100 |
[64] |
N, the number of subjects
SNP (c.-617C>A) in the Nrf2 gene as a biomarker for prognosis of lung cancer
The Nrf2 gene is regarded as a double-edged sword. It plays a key role in protecting normal cells from oxidative stress as well as external toxic challenges, whereas it can also endow cancer cells resistance to anticancer drugs. Recently it has been reported that Nrf2 contributes to malignant phenotypes of cancer cells in vitro, including aggressive cell proliferation, drug resistance, and metabolic re-programming [46,47]. Indeed, Nrf2 activation and/or up-regulated expression of the Nrf2 gene are involved in the emergence of cancer resistance to various anticancer drugs by transcriptionally activating a battery of cell-protection genes [48,49].
The SNP -617C>A [43] is considered to regulate the positive feedback loop of transcriptional activation of the Nrf2 gene and thereby it can modulate the Nrf2 protein expression level (Figure 4). Since the SNP (-617A) in the ARE-like loci of the human Nrf2 gene decreases the binding affinity to the transcription factors of Nrf2/small Maf (MafK) [44], it is anticipated that the homozygous -617 A/A allele significantly attenuate the positive feedback loop of transcriptional activation of the Nrf2 gene.
The authors have recently found that lung cancer patients with homozygous SNP alleles (-617A/A) in the Nrf2 gene had markedly high overall survival [50]. Univariate analysis showed a significant difference between the -617 A/A and -617 C/A groups in terms of overall survival (log-rank P = 0.021). As one of the intrinsic genetic polymorphisms in the Nrf2 gene, homozygous SNP (-617A/A) is considered to be associated with overall survival of lung cancer patients [50].
Nrf2 is known to regulate the basal expression of the murine double minute-2 (MDM2) gene [51]. Since human MDM2 is an oncoprotein that binds to p53 protein and inactivates the tumor suppressor activity of p53 [52], Nrf2 can indirectly contribute to p53-mediated cell cycle control and/or apoptosis [53]. This, in turn, attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans [54]. In addition, Nrf2 activation may be involved in the emergence of cancer resistance to various anticancer drugs by transcriptionally activating a battery of self-defense genes. For example, ABCG2 is one of the Nrf2 target genes and it is known to mediate the efflux of gefitinib from cancer cells [55]. Its expression is regulated by Nrf2 [56] and the EGFR-tyrosine kinase cascade [57,58]. In this context, it is suggested that the genetic polymorphism in the Nrf2 gene could be a clinically useful biomarker for prognosis of lung cancer patients.
Concluding remarks
Recently, Figarska et al. genotyped SNPs, i.e., rs4243387, rs2364723, rs13001694, rs1806649, and rs6726395) located in intron 1 between exons 1 an2 of the human Nrf2 gene in 1,390 subjects from the Vlagtwedde-Vlaardingen cohort in the Netherlands [59]. Their study suggested that Nrf2 is indeed associated with reduced risk of cardiovascular and COPD mortality. At present, however, information is still limited as to the clinical impact of genetic polymorphisms of the Nrf2 gene. Insight into molecular mechanisms underlying the impact of genetic polymorphisms as well as epidemiological studies with genotyping should be necessary. In addition to the above-mentioned genetic polymorphisms as the “intrinsic” mechanism, mutations in the Keap1 and/or Nrf2 genes are the “acquired” mechanisms that lead to constitutive activation of Nrf2. In fact, mutations in the Nrf2 and Kea1 genes have been found in carcinomas of the lung [60], breast [61], liver [62], and stomach [62]. Abnormalities in Nrf2 activity were correlated with poor prognosis, when measured either as recurrence-free or overall 5-year survival. A recent immunohistochemical study has revealed that increased expression of Nrf2 protein and decreased expression of Keap1 protein are common abnormalities in non-small cell lung cancer (NSCLC) and are associated with poor prognosis [50]. Importantly, abnormal expression of Nrf2 and Keap1 proteins was more common than that of the corresponding gene mutaions [63], suggesting the involvement of other mechanisms such as intrinsic genetic polymorphisms of those genes. Genetic polymorphisms/mutations and fine balances among Nrf2, Keap1, MDM2, p53, p21WAF1/cip1 and other genes are likely to contribute to the progression of cancer and, consequently, the prognosis of cancer patients.
Conflict of interest/disclosure
Hereby the authors declare that there is no conflict of interest for the present study.
References
2021 Copyright OAT. All rights reserv
- Manhesa G, Allègre CJ, Dupréa B, Hamelin B (1980) Lead isotope study of basic-ultrabasic layered complexes: Speculations about the age of the earth and primitive mantle characteristics. Earth Planet Sci Lett 47: 370–382
- Noffke, N, Christian D, Wacey D, Hazen RM (2013) Microbially Induced Sedimentary Structures Recording an Ancient Ecosystem in the ca. 3.48 Billion-Year-Old Dresser Formation, Pilbara, Western Australia. Astrobiol 13: 1103–1124. [Crossref]
- Ohtomo Y, Kakegawa T, Ishida A. et al. (2014) Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks. Nat Geoscie 7: 25–28.
- Bell EA, Boehnke P, Harrison TM, Mao WL (2015) Potentially biogenic carbon preserved in a 4.1 billion-year-old zircon. Proc Natl Acad Sci U S A 112: 14518-14521. [Crossref]
- Schopf JW (2006) Fossil evidence of Archaean life. Philos Trans R Soc Lond B Biol Sci 361: 869-885. [Crossref]
- Finkel T (2003) Oxidant signals and oxidative stress. Curr Opin Cell Biol 15: 247-254. [Crossref]
- Meister A, Anderson ME (1983) Glutathione. Annu Rev Biochem 52: 711-760. [Crossref]
- Winyard PG, Moody CJ, Jacob C (2005) Oxidative activation of antioxidant defence. Trends Biochem Sci 30: 453-461. [Crossref]
- Dröge W (2002) Free radicals in the physiological control of cell function. Physiol Rev 82: 47-95. [Crossref]
- Reed DJ (1990) Glutathione: toxicological implications. Annu Rev Pharmacol Toxicol 30: 603-31. [Crossref]
- Fahey RC, Sundquist AR (1991) Evolution of glutathione metabolism. Adv Enzymol Relat Areas Mol Biol 64: 1-53. [Crossref]
- Sies H (editor) (1985) Oxidative stress. Orlando: Academic Press.
- Rahman I, Adcock IM (2006) Oxidative stress and redox regulation of lung inflammation in COPD. Eur Respir J 28: 219-242. [Crossref]
- Shinohara T, Kaneko T, Nagashima Y, Ueda A, Tagawa A, Ishigatsubo Y (2005) Adenovirus-mediated transfer and overexpression of heme oxygenase 1 cDNA in lungs attenuates elastase-induced pulmonary emphysema in mice. Hum Gene Ther 16: 318-327. [Crossref]
- Sato T, Takeno M, Honma K, Yamauchi H, Saito Y, et al. (2006) Heme oxygenase-, a potential biomarker of chronic silicosis, attenuates silica-induced lung injury. Am J Respir Crit Care Med 174: 906-914. [Crossref]
- Mishina K, Shinkai M2, Shimokawaji T, Nagashima A, Hashimoto Y, et al. (2015) HO-1 inhibits IL-13-induced goblet cell hyperplasia associated with CLCA1 suppression in normal human bronchial epithelial cells. Int Immunopharmacol 29: 448-453. [Crossref]
- Moi P, Chan K, Asunis I, Cao A, Kan YW (1994) Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 91: 9926-9930. [Crossref]
- Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M (1995) Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol 15: 4184-4193. [Crossref]
- Motohashi H, Yamamoto M (2004) Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med 10: 549-557. [Crossref]
- Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB (2005) Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem 280:32485-32492. [Crossref]
- Loboda A, Damulewicz M, Pyza E, Jozkowicz A, et al. (2016) Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci. [Crossref]
- Kobayashi M, Yamamoto M (2006) Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul 46: 113-140. [Crossref]
- Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, et al. (2008) Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28: 2758-2770. [Crossref]
- Martin D, Rojo AI, Salinas M, Diaz R, Gallardo G, et al. (2004) Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J Biol Chem 279: 8919-8929. [Crossref]
- Kocanova S, Buytaert E, Matroule JY, Piette J, Golab J, et al. (2007) Induction of heme-oxygenase 1 requires the p38MAPK and PI3K pathways and suppresses apoptotic cell death following hypericin-mediated photodynamic therapy. Apoptosis 12:731-741. [Crossref]
- Andreadi CK, Howells LM, Atherfold PA, Manson MM (2006) Involvement of Nrf2, p38, B-Raf, and nuclear factor-kappaB, but not phosphatidylinositol 3-kinase, in induction of hemeoxygenase-1 by dietary polyphenols. Mol Pharmacol 69: 1033-1040. [Crossref]
- Bloom DA, Jaiswal AK (2003) Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem 278:44675-44682. [Crossref]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, et al. (2003) Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23: 7198-7209. [Crossref]
- Cullinan SB, Diehl JA (2004) PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279: 20108-20117. [Crossref]
- Kang KW, Lee SJ, Park JW, Kim SG (2002) Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol Pharmacol 62: 1001-1010. [Crossref]
- Kang KA, Lee KH, Park JW, Lee NH, Na HK, et al. (2007) Triphlorethol-A induces heme oxygenase-1 via activation of ERK and NF-E2 related factor 2 transcription factor. FEBS Lett 581: 2000-2008. [Crossref]
- Igarashi K, Sun J (2006) The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal 8: 107-118. [Crossref]
- Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, et al. (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16: 6083-6095. [Crossref]
- Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, et al. (2002) Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J 21: 5216-5224. [Crossref]
- Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, et al. (2004) Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci U S A 101: 1461-1466. [Crossref]
- Hintze KJ, Katoh Y, Igarashi K, Theil EC (2007) Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase, beta-globin, and NADP(H) quinone (oxido) reductase1. J Biol Chem 282: 34365-34371. [Crossref]
- Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, et al. (2003) Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J Biol Chem 278: 9125-9133. [Crossref]
- Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL (2006) Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. FASEB J 20: 2651-2653. [Crossref]
- Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, et al. (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J 20: 2835-2843. [Crossref]
- Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, et al. (2004) Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J 23: 2544-2553. [Crossref]
- Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, et al. (2007) Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol 27: 6962-6971. [Crossref]
- Reichard JF, Motz GT, Puga A (2007) Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res 35: 7074-7086. [Crossref]
- Yamamoto T, Yoh K, Kobayashi A, Ishii Y, Kure S, et al. (2004) Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun 321: 72-79. [Crossref]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, et al. (2007) Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21: 2237-2246. [Crossref]
- Ishikawa T, Toyoda Y, Yoshiura K, Niikawa N (2013) Pharmacogenetics of human ABC transporter ABCC11: new insights into apocrine gland growth and metabolite secretion. Front Genet 3: 306. [Crossref]
- Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, et al. (2009) Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res 15: 3423-3432. [Crossref]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, et al. (2012) Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66-79. [Crossref]
- Nakata K, Tanaka Y, Nakano T, Adachi T, Tanaka H, et al. (2006) Nuclear receptor-mediated transcription regulation in phase I, II, and III xenobiotic metabolizing systems. Drug Metab Pharmacokinet 21:437-457.
- Shen G, Kong AN (2009) Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos 30: 345-355. [Crossref]
- Okano Y, Nezu U, Enokida Y, Lee MT, Kinoshita H, et al. (2013) SNP (-617C>A) in ARE-like loci of the NRF2 gene: a new biomarker for prognosis of lung adenocarcinoma in Japanese non-smoking women. PLoS One 8: e73794. [Crossref]
- You A, Nam CW, Wakabayashi N, Yamamoto M, Kensler TW, et al. (2011) Transcription factor Nrf2 maintains the basal expression of Mdm2: An implication of the regulation of p53 signaling by Nrf2. Arch Biochem Biophys 507: 356-364. [Crossref]
- Freedman DA, Wu L, Levine AJ (1999) Functions of the MDM2 oncoprotein. Cell Mol Life Sci 55: 96-107. [Crossref]
- Rotblat B, Melino G, Knight RA (2012) NRF2 and p53: Januses in cancer? Oncotarget 3: 1272-1283. [Crossref]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, et al. (2004) A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591-602. [Crossref]
- Saito H, Hirano H, Nakagawa H, Fukami T, Oosumi K, et al. (2006) A new strategy of high-speed screening and quantitative structure-activity relationship analysis to evaluate human ATP-binding cassette transporter ABCG2-drug interactions. J Pharmacol Exp Ther 317: 1114–1124. [Crossref]
- Singh A, Wu H, Zhang P, Happel C, Ma J, et al. (2010) Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Mol Cancer Ther 9: 2365-2376. [Crossref]
- Meyer zu Schwabedissen HE, Grube M, Dreisbach A, Jedlitschky G, Meissner K, et al. (2006) Epidermal growth factor-mediated activation of the map kinase cascade results in altered expression and function of ABCG2 (BCRP). Drug Metab Dispos 34: 524–533. [Crossref]
- Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, et al. (2011) Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem 286: 20558–20568. [Crossref]
- Figarska SM, Vonk JM, Boezen HM (2014) NFE2L2 polymorphisms, mortality, and metabolism in the general population. Physiol Genomics 46: 411-417. [Crossref]
- Sporn MB, Liby KT (2012) NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564-571. [Crossref]
- Sjӧblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. [Crossref]
- Yoo NJ, Kim HR, Kim YR, An CH, Lee SH (2012) Somatic mutations of the KEAP1 gene in common solid cancers. Histopathology 60: 943-952. [Crossref]
- Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, et al. (2010) Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 16: 3743-3753. [Crossref]
- http://browser.1000genomes.org/Homo_sapiens/Variation/Population?db=core;r=2:178129537-178130537;v=rs6721961;vdb=variation;vf=4574214





