Background: There is recent concern about the development of colistin resistance that may disturb the antibiotics therapy used for extended beta lactamase producing Enterobacteriaceae species.
Aim: The aim of the present study was to investigate the presence of mcr-1 and mcr-2 genes in clinical isolates of Enterobacteriaceae spp. resistant to colistin.
Study design: Retrospective laboratory based study.
Material and Method The study was conducted on 50 Enterobacteriaceae species resistant to colistin collected from clinical samples from patients with health care associated infections according to CDC definitions. Minimum inhibitory concentrations (MICs) of colistin was performed by the use of the broth microdilution method according to CLSI. Isolates were reported resistant if MIC was >2 mg/L. Polymerase chain reaction (PCR) for mcr-1 and mcr-2 was performed.
Results: Colisitin resistance genes was detected by PCR in 2 isolates. Mcr-1 gene was detected in 2 isolates (4%) and mcr-2 was not detected in any isolates. Mcr-1 was detected in one E.coli strain and in one K.pneumoniae strains. The presence of Mcr-1 was associated with in high MIC >16mg/L.
Conclusion: The present study highlights the emergence of colistin resistance among E.coli and K.pneumoniae in tertiary health care setting. The gene that was responsible for this resistance was mcr-1 while mcr-2 was not detected. There is a need for future studies with large number of clinical isolates to determine the prevalence of colistin resistance and the responsible molecular mechanism for such resistance.
Colistin resistance, Enterobacteriaceae, E. coli, Klebsiella pneumonia, PCR, mcr-1, mcr-2
There is worldwide increase in the prevalence of antibiotics resistance among Enterobacteriaceae species (spp.). The resistance to antibiotics results in increase health care costs with increase in morbidity and mortality rates among patients especially in health care related infections [1,2]. Resistance among Enterobacteriaceae species to β-lactam antibiotics leaves few therapeutic options to be used among which are carbapenem antibiotics and polymyxin B. Unfortunately, there are several evidence of wide spread of carbapenem resistance [3,4]. This emergence of resistance, makes the polymyxin B the last resort for treatment of infections with carbapenemase producing Enterobacteriaceae. This finding, leads WHO to classify colistin as an important for human medicine [5].
The mode of action of colistin depends upon the interaction of it with the outer membrane of the lipopolysacharide portion of the bacterial cell membrane leading to its lyses. The resistance to colistin arises from two mechanisms either chromosomal mutations or plasmid acquiring resistance [6]. The chromosomal mutations occur in the genes encoding the PmrA/PmrB and PhoP/PhoQ, the negative regulator MgrB leading either to the modifications or even loss of the lipid A molecule. This mutations are associated with the use of colistin [7]. The other type of resistance is plasmid mediated resistance to colistin that confers to the presence of mcr-1 gene which encodes for a phosphoethanolamine transferase enzyme that leads to transfers a phosphoethanolamine to Lipid A; conferring resistance to colistin. Another gene mcr-2 was also identified to be associated with colistin resistance [8]. The plasmid mediated resistance is stable resistance, not related to the use of colistin and it is found essentially in E. coli, Pseudomonas aeruginosa and Klebsiella pneumoniae [6]. Further studies have reported the presence of the mcr- 1 gene in different Enterobacteriaceae species beside E. coli, Klebsiella pneumonia (K. pneumonia) such as Salmonella spp., in different geographical regions including Asia, Europe, North America and Africa [9-17]. The main risk for plasmid mediated resistance is the easy transfer between different gram negative species. If this occurs to species already resistance to carbapenem antibiotics in health care associated infections, this will main the presence of non treated infections [18].
The aim of the present study was to investigate the presence of mcr-1 and mcr-2 genes in clinical isolates of Enterobacteriaceae spp. resistant to colistin from health-care associated infections in Mansoura University hospitals.
The study was conducted on 50 Enterobacteriaceae species resistant to colistin collected from clinical samples from patients with health care associated infections according to CDC. The study was approved by the ethical committee of our institute. Microbiological identifications of the isolates were performed by automated Microscan system (Beckman Coulter International, USA). Antibiotics susceptibility was performed by disc diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [19] to ceftazidime, cefotaxime, cefepime, imipenem, meropenem, gentamicin, amikacin, ciprofloxacin, levofloxacin, sulfamethoxazole/trimethoprim and piperacillin/tazobactam. In addition, minimum inhibitory concentrations (MICs) of colistin was performed by the use of the broth microdilution method according to CLSI. Isolates were reported resistant if MIC was >2 mg/L [19].
DNA extraction
Pure colonies of isolates were cultured on nutrient broth at 37C for 24 hours. Later on, 100 micron of broth was centrifuged for 5 minutes and the deposit was resuspended in 100 micron distilled sterile water and heated in water bath at 95C for 20 minutes. The supernatant was collected in sterile eppendorf and kept frozen at -20C till amplification.
Amplification and detection of mcr-1 and mcr-2
The sequences of the used primers were summarized in Table 1, [8,20]. For amplification Qiagen amplification master mix was used (Qiagen). Total amplification volume was 25 micron with 3 μl of the bacterial crude lysate and 0.5 μM of each primer. The amplification procedure was performed with the following steps 5 min at 94°C, followed by 30 cycles of 45 s at 94°C, 1 min at 60°C (for mcr-1) or 1 min at 55°C (for mcr-2), 1 min at 72°C, and a final extension time of 7 min at 72°C [8,10,21].
Table 1. Genes and primers sequences with amplified bp
Gene |
Sequence |
bp |
mcr-1 |
F:5/-CGGTCAGTCCGTTTGTTC-/3
R:5/-CTTGGTCGGTCTGTA GGG-/3 |
309 bp |
mcr-2 gene |
F: 5′ TGGTACAGCCCCTTTATT 3′
R: 5′ GCTTGAGATTGGGTTATGA 3′ |
567 bp
|
Electrophoresis with gel 2% was performed for 20 minutes. The products was visualized by UV and compared with DNA ladder. The amplified products was confirmed by sequence analysis.
The clinical isolates of Enterobactericae spp. resistant to colistin was collected during 36 months from Mansoura University hospitals. The most common sources were urine (46%), blood (30%) and wounds (24%), Table 2.
Table 2. Source of colistin resistant strains
Source |
No. |
% |
blood |
15 |
30.0 |
urine |
23 |
46.0 |
wound |
12 |
24.0 |
Total |
50 |
100.0 |
The isolates were K.pneumonia (44%), E.coli (42%), Enterobacter species (10%) and Acinetobacter baumannii (4%), Table 3.
Table 3. Isolated Enterobactericae species
Enterobactericae species |
No. |
% |
A. baumanni |
2 |
4.0 |
E. coli |
21 |
42.0 |
Enterobacter |
5 |
10.0 |
K. pneumoniae |
22 |
44.0 |
Total |
50.0 |
100.0 |
The isolated strains had marked resistance to the third generation cephalosporines ceftazidime (60%) and cefotaxime (56%) and fourth generation cephalosporine, cefepime (78%). Resistance to carbapenem antibiotics imipenem and meropenem was 50% and 44% respectively. Less resistance was noticed for amikacin 42% and gentamicin (40%), Table 4.
Table 4. Antibiotics resistance of the isolated Enterobactericae spp
Antibiotics |
No. % |
Amikacin |
21 42% |
cefepime |
39 78% |
cefotaxime |
28 56% |
ceftazidime |
30 60% |
imipenem |
25 50% |
meropenem |
22 44% |
levofloxacin |
33 66% |
gentamicin |
20 40% |
ciprofloxacin |
32 64% |
sulfamethoxazole/trimethoprim |
22 44% |
piperacillin/tazobactam |
33 66% |
Minimal inhibitory concentrations for colistin was found to be >16mg/L in 30 isolates (60%) , 8-16mg/L in 8 isolates (16%) and 4-8 mg/L in 12 isolates (24%), Table 5, Figure 1.
Table 5. Distribution of levels of minimal inhibitory concentration of colistin among isolated strains
MIC Mg/L |
No. |
% |
>16 |
30 |
60 |
Aug-16 |
8 |
16 |
04-Aug |
12 |
24 |
Total |
50 |
100 |
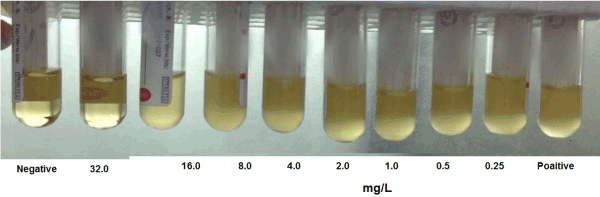
Figure 1. MICS of resistant isolate to colistin Negative: Negative control broth without culture Positive: Positive control broth with culture without colistin.
Colisitin resistance genes was detected by PCR in 2 isolates. Mcr-1 gene was detected in 2 isolates (4%) and mcr-2 was not detected in any isolates, Table 6.
Table 6. Frequency of Mcr-1 and Mcr-2 detection by PCR
Gene |
No. |
% |
Mcr-1 |
2 |
4 |
Mcr-2 |
0 |
0 |
Total |
50 |
100 |
Mcr-1 was detected in one E.coli strain and in one K.pneumoniae strains, Table 7.
Table 7. Distribution of Mcr-1 among Enterobactericae species
Enterobactericae species |
Mcr-1 No. (%) |
N0. (%) |
E. coli |
1 (4.7%) |
21 (100%) |
K. pneumoniae |
1 (4.5%) |
22 (100%) |
The presence of Mcr-1 was associated with in high MIC >16mg/L, Table 8, Figure 2.
Table 8. Distribution of Mcr-1 according to MICs of colistin.
MIC of colistin |
Mcr-1 |
N0. (%) |
No. (%) |
>16 |
2 3.3% |
30 100% |

Figure 2. Positive PCR of the two isolates for mcr-1 M: marker Lanes 1, 2 positive isolates.
The emergence of colistin resistance among clinical isolates of Enterobactericae spp. Has been reported in recent years due to increase its use for carbapenem resistant isolates [3,4]. The association of colistin resistance by plasmid that can be transferred between different gram negative bacilli leads to arise of other danger in the era of antibiotics resistance and making the study of the presence of colistin resistance an important issue to limit its spread [5].
In the present study, 50 colistin resistant isolates during 36 months had been collected in which the most common sources were urine (46%), blood (30%) and wounds (24%).Similar result was obtained by previous study [22-24].
The isolates were K.pneumonia (44%), E.coli (42%), Enterobacter species (10%) and Acinetobacter baumannii (4%). Previous reports were online with these results as K. Pneumoniae, E. coli A. baumannii and Enterobacter cloacae were frequently associated with colistin resistance [23-25].
The association of these isolates with colistin resistance may be associated with the previous use of colistin [25], or even without previous colistin therapy [26].
The isolated strains had marked resistance to the third generation cephalosporines ceftazidime (60%) and cefotaxime (56%) and fourth generation cephalosporine, cefepime (78%). Around half of the isolates had resistance to carbapenem antibiotics.
Several reports had documented the association of carbapenem resistance and resistance to the third generation of cephalosporines with colistin resistance among different species of Enterobactericaea [27-30]. Therefore there is a need for the study of the prevalence of colistin resistance among isolates with extended spectrum beta lacatamase resistance [31,32]. Less resistance was noticed for amikacin 42% and gentamicin (40%). This on contrary to previous results reporting poor sensitivity to aminoglycosides [23,33]. This difference in susceptibility may reflect the difference of antibiotics policy among different health care settings in different geographical regions.
The use of PCR to detect mcr-1 gene in the present study has revealed the presence of mcr-1 in one E.coli strain and in one K.pneumoniae strain. There are several reports about the dissemination of strains of K.pneumoniae and E.coli with detected mcr-1 gene including Egypt and Arabian Peninsula [34,35]. These findings highlights that the presence of such gene among clinical isolates. Thus the presence of mcr-1 should be monitored and studies should be carried on large number of isolates. Spread of the mcr-1 gene in the community and successively in the hospital would pose a threat to patients developing an infection with mcr-1 containing multidrug resistant Enterobacteriaceae isolates as this will limit the therapeutics options [36].
In the present study none of the isolates had mcr-2. Recently, it was reported that a novel gene carried on plasmid-, mcr-2, also confers resistance to colistin [8], although it seems unusual that the mcr-2 gene is detected only in Belgium [37]. This posed a hypothesis that might be due to a mechanism for mcr-2 dissemination different from that of the paradigm mcr-1 gene.
The presence of Mcr-1 was associated with in high MIC >16mg/L. Generally most isolates with colistin resistant strains had MICs in the range of 4 or 8 mg/L. Even in one study the strains which harbor mcr-1 were susceptible to colistin with a MIC of ≤ 0.25mg/l. The discrepancy in the results of MICs may be attributed to the difference in the number of the tested isolates.
The present study highlights the emergence of colistin resistance among E.coli and K.pneumoniae in tertiary health care setting. The gene that was responsible for this resistance was mcr-1 while mcr-2 was not detected. There is a need for future studies with large number of clinical isolates to determine the prevalence of colistin resistance and the responsible molecular mechanism for such resistance.
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States (2013).
- World Health Organization (2014) Antimicrobi2021 Copyright OAT. All rights reservillance.
- Livermore DM, Woodford N (2006) The beta lactamase threat in Enterobacteriaceae, pseudomonas and Acinetobacter. Trends Microbiol 14: 413-420. [Crossref]
- Walsh TR (2010) Emerging carbapenemases: a global perspective. Int J Antimicrob Agents 36: S8–S14. [Crossref]
- World Health Organization (2016) Critically important antimicrobials for human medicine.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16: 161-168. [Crossref]
- Olaitan AO, Morand S, Rolain JM (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5: 1-18. [Crossref]
- Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, et al. (2016) Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21. [Crossref]
- Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, et al. (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 144-145. [Crossref]
- Tse H, Yuen KY (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 145-146. [Crossref]
- Hu Y, Liu F, Lin IY, Gao GF, Zhu B (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 146-147. [Crossref]
- Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 147. [Crossref]
- Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, et al. (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16: 147-149.
- Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, et al. (2016) Occurrence of plasmid-borne mcr-1 Colistin resistance gene in extendedSpectrum-β-Lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother 60: 2594–2595. [Crossref]
- Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, et al. (2016) Detection of plasmid-mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci 105: 134-135. [Crossref]
- Elnahriry SS, Khalifa HO, Soliman AM, Ahmed AM, Hussein AM, et al. (2016) Emergence of plasmid-mediated Colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother 60: 3249-3250. [Crossref]
- Coetzee J, Corcoran C, Prentice E, Moodley M, Mendelson M, et al. (2016) Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J 106: 35-36. [Crossref]
- Newton-Foot M, Snyman Y, Maloba MRB, Whitelaw AC (2016) Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control 6: 78.
- Clinical and Laboratory Standards Institute (CLSI) M100-S23 (2013) Performance standards for antimicrobial susceptibility testing; twenty third informational supplement.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16: 161-168. [Crossref]
- Quiñones B, Swimley MS, Narm KE, Patel RN, Cooley MB, et al. (2012) O-antigen and
virulence profiling of Shiga toxin-producing Escherichia coli by a rapid and cost-effective DNA
microarray colorimetric method. Front Cell Infect Microbiol 2: 61. [Crossref]
- Arjun R, Gopalakrishnan R, Nambi PS, Kumar DS, Madhumitha R, et al. (2017) A Study of 24 Patients with Colistin-Resistant Gram-negative Isolates in a Tertiary Care Hospital in South India. Indian J Crit Care Med 21: 317-321. [Crossref]
- Goel G, Hmar L, Sarkar De M, Bhattacharya S, Chandy M (2014) Colistin-resistant Klebsiella pneumoniae: Report of a cluster of 24 cases from a new oncology center in Eastern India. Infect Control Hosp Epidemiol 35: 1076-1077. [Crossref]
- Ramesh N, Gothandam KM, Prasanth M, Ramkumar S, Karthikeyan S, et al. (2016) Colistin susceptibility of gram-negative clinical isolates from Tamil Nadu, India. Asian Biomed 10: 35-39.
- Qureshi ZA, Hittle LE, O’Hara JA, Rivera JI, Syed A, et al. (2015) Colistin-resistant Acinetobacter baumannii: Beyond carbapenem resistance. Clin Infect Dis 60: 1295-1303. [Crossref]
- Chen S, Hu F, Zhang X, Xu X, Liu Y, et al. (2011) Independent emergence of colistin-resistant Enterobacteriaceae clinical isolates without colistin treatment. J Clin Microbiol 49: 4022-4023. [Crossref]
- Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, et al. (2014) Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: A rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill 19: 20939. [Crossref]
- Prim N, Rivera A, Rodriguez-Navarro J, Espanol M, Turbau M, et al. (2016) Detection of mcr-1 colistin resistance gene in polyclonal Escherichia coli isolates in Barcelona, Spain, 2012 to 2015. Euro Surveill 21(13). [Crossref]
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Hoang HT, et al. (2016) Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis 16: 286-287. [Crossref]
- Ghafur A, Vidyalakshmi PR, Murali A, Priyadarshini K, Thirunarayan MA (2014) Emergence of pan-drug resistance amongst gram negative bacteria! The first case series from India. J Microbiol Infect Dis 4: 86-91.
- Du H, Chen L, Tang YW, Kreiswirth BN (2016) Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287-278. [Crossref]
- Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW (2016) Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 16: 285-286. [Crossref]
- Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, et al. (2016) First Description of mcr-1- Mediated Colistin Resistance in Human InfectionsCaused by Escherichia coli in Latin America. Antimicrob Agents Chemother 60: 4412-4413. [Crossref]
- Sonnevend Á, Ghazawi A, Alqahtani M, Shibl A, Jamal W, et al. (2016) Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis 50: 85-90. [Crossref]
- Terveer EM, Nijhuis RHT, Crobach MJT, Knetsch CW, Veldkamp KE, et al. (2017) Prevalence of colistin resistance gene (mcr1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1containing, colistin susceptible E. coli. PLoS One 12: e0178598. [Crossref]
- Liassine N, Assouvie L, Descombes MC, Tendon VD, Kieffer N, et al. (2016) Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int J Infect Dis 51:4-5. [Crossref]
- Sun J, Xu Y, Gao R, Lin J, Wei W, et al. (2017) Deciphering MCR-2 Colistin Resistance. MBio. 8. [Crossref]


