Traumatic brain injury (TBI) is associated with a collection of physical, emotional and cognitive post complications. The background for such complex consequences may be due to a number of different factors, and the nature of these changes indicates that the frontal lobes may be implicated. In this study we have employed gene expression arrays and gene ontology databases to search for possible similarities between different forms of acquired brain injuries in order to test whether molecular relationships exist between the different pathologies. Two types of experimental models for traumatic brain injuries, lateral fluid percussion and rotational acceleration, were used. Their molecular signature was identified and compared with those related to other rodent models simulating stress, depression, alcohol dependence, stroke, and Alzheimer’s disease. The data show that the two TBI models share similar gene expression changes with the models with regard to depression and stroke, indicating a common molecular support between these pathologies. The data provided can contribute to the introduction of new treatment strategies related to TBI.
frontal lobe, microarray, functional network, lateral fluid percussion trauma, rotational trauma
Traumatic brain injury (TBI) can have many different origins, such as motor vehicle accidents, falls, sport activities, assaults or, blast waves from explosions. Brain damage following TBI may result in an increased risk of developing secondary pathological conditions, including mental health disorders and neurodegenerative diseases [1-4]. Mental health disorders following TBI include stress, anxiety, depression, and substance abuse, such as alcohol dependence, and it is becoming increasingly clear that TBI can potentially progress into a degenerative disorder affecting the brain and other organs [5,6]. In particular, accumulation of amyloid deposition, a biomarker of Alzheimer’s disease, has been described following TBI in humans [7] and animal models [8,9]. TBI has also been recognized as a potential stroke risk factor due to vascular dissection or abnormal coagulation [10] while TBI and ischemic stroke have been shown to share common secondary insults such as an increase in neurotransmitter levels, neuronal apoptosis, and immune-inflammatory activation in particular. Overall, TBI is associated with a collection of physical, emotional and cognitive post complications, but to date there are not any convincing theories for understanding why these complications appear after brain trauma. Consequently, the question as to what relationships exist between TBI and such secondary pathological conditions still remains. A better understanding of the underlying brain molecular mechanisms could help to solve this issue.
Several experimental animal models have been designed to model brain trauma [11]. In the present study, two rat models were investigated: lateral fluid percussion (lfp-TBI) and rotational acceleration (rot-TBI). The lfp-TBI produces a brief deformation of the brain tissue near to the trauma device attachment point. The trauma induces intracranial haemorrhage, brain swelling and progressive grey matter damage, all of which are pathophysiological consequences of TBI in humans [12-14]. The rot-TBI mimics a forehead impacting a stiff surface which is common in traffic, sports and fall accidents. This model mainly produces distributed axonal injuries [15].
Gene expression changes in the two rat TBI models were studied at a post-trauma time of 1 month. The reasoning behind imposing such an extended post-injury period was to ensure that mainly secondary or even tertiary events in the molecular signature of traumatized brains were included, and not genes associated with acute primary insult [16-21].
The frontal cortex was examined as it is known to be involved in important functions such as attention, behavioral flexibility, and working memory. The frontal cortex receives and integrates information from multiple brain regions, including the hippocampus, and many neuromodulators target its functions [22]. In addition, it was included as functional neuroimaging has consistently shown abnormalities in prefrontal regions in mild TBI patients [23,24].
The aim of the present study was to identify the molecular signature of traumatized brains (lfp- and rot-TBI) and to compare them with published case studies on stress, depression, alcohol dependence, stroke, and Alzheimer’s disease in order to establish if a common molecular signature would provide evidence of the molecular nature of the relationship between TBI and the above mentioned post complications. Establishing a relationship has the potential for major therapeutic consequences as specific medication addressing all post-TBI challenges is not available as yet [4]. Subsequently, identification of specific molecules involved in both TBI and secondary pathologies would likely offer a new direction for treatment strategies.
Experimental brain injury models
Lateral fluid percussion brain injury rat model: Eight adult male Sprague–Dawley rats (250–300 g; Harlan, France) were anaesthetized with a mix of ketamine (Imalgen® 1000; 800 µl/kg) and xylazine (Rompun 2%; 500 µl/kg). Ropivacain (Naropeine TM) was injected subcutaneously before scalp incision. After scalp incision, a 3 mm diameter craniotomy was created and centred at -3 mm from bregma and 3.8 mm left of the sagittal suture. One steel screw was inserted into the skull, 1 mm rostral to bregma. A female Luer-Loc hub (inside diameter of 4.0 mm) was centred over the craniotomy site and bonded to the skull with cyanoacrylate glue, it was then filled with sterile saline. Dental acrylic (Paladadur®, Heraeus Kulzer GmbH, Germany) was used to bond the Luer hub to the skull. Post-surgery, the animals were returned to their cages. Four and a half hours post the end of the surgical procedure; animals were re-anaesthetized with isoflurane and then connected to the Picospritzer-Fluid Percussion Injury apparatus. The Picospritzer solenoid was triggered and a pressure wave was applied to the brain cavity. Peak pressure and pressure duration values were measured offline. Once removed from the apparatus, the animals were returned to their home cages. Sham animals were subjected to anaesthesia and surgery but not to trauma. All surgical procedures were approved by the University Claude Bernard Institutional Committee for Animal Care and Use in Lyon, France.
Rotational brain injury rat model: Eight adult male Sprague–Dawley rats (460 – 570 g; Taconic, Denmark) were anaesthetized with 2.4 ml/kg intra-abdominal injections of a mixture of 1ml Dormicum® (5 mg/ml Midazolan, Roche), 1 ml Hypnorm® (Janssen) and 2 ml of distilled water. A midline incision was made through the skin and periosteum of the skull vault, and parts of the frontal, nose and parietal bones were freed from adherent tissue. The exposed bone was treated with 15% phosphate acid for 3 minutes, thoroughly rinsed with tempered saline, and dried for 3 minutes with an air dryer providing air at 37°C in the proximity of the skull bone. The exposed bone was then gently sanded and dried for at least an additional 10 minutes, prior to gluing of a curved aluminium plate, denominated the skull cap, and shaped to match the contour of the exposed skull. The glue (Super-Bond C&B dental glue; Sun Medical Co., Japan) was left to cure for a minimum of 15 minutes at approximately 37°C. Post-surgery, an attachment plate was fastened by means of two screws to the skull cap previously glued to the skull bone. Then the attachment plate was inserted and secured to a rotating bar that was able to rotate freely around a horizontal axis perpendicular to the animal sagittal plane. The resulting pre-trauma position of the head was slightly flexed and the brain centre of gravity located about 6.5 mm above the centre of rotation. During trauma a solid brass weight was accelerated in a specially designed air driven accelerator (air pressure 14 bar, speed 50.9 ± 0.07 m/s) and was made to hit a rubber block glued onto a striker target (for additional details see Davidsson and Risling, 2011) [25]. The impulse produced subjected the rotating bar and the animal head to a short sagittal plane rearward rotational acceleration (1.75 ± 0.13 Mrad/s2). Post trauma the attachment plate was removed, the skin was made to cover the skull cap and 8 to 10 sutures closed the incision. Sham animals were subjected to anaesthesia and surgery but not to trauma. All surgical procedures were approved by the Animal Care and Use Ethics Committee in Stockholm, Sweden.
Tissue collection and RNAs extraction: Left frontal cortices (N=16) were dissected from the bregma level of 3.70 mm in accordance with Paxinos and Watson stereotaxic atlas (1998) [26]. Particular attention was paid to not include the olfactory lobe. Tissues were then homogenized and purified with a mirVana miRNA Kit (AB Applied-Ambion, France) according to the manufacturer’s protocol. The quality and quantity of total Ribonucleic Acid (RNA) was determined with an Agilent 2100 Bioanalyzer (Agilent Technologies, France). The samples were selected for microarray or for quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) experiments provided that they had an >8.0 RNA integrity number, a clear gel image and no Deoxyribonucleic Acid (DNA) contamination as observed on the histogram.
Microarray assay and analysis: Sample amplification, labelling, and hybridization followed the one-color microarray-based gene expression analysis (low input quick amp labelling) protocol (version 6.5, May 2010) recommended by Agilent Technologies. Thereby, 200 ng of each total RNA sample was reverse transcribed into complementary DNA (cDNA) using oligo dT-T7 promoter primer. Labelled complementary RNA (cRNA) was synthesized from the cDNA. The reaction was performed in a solution containing a deoxyuridine triphosphate (dNTP) mix, cyanine 3-dCTP, and T7 RNA Polymerase, and incubated at 40°C for 2 hrs. Qiagen’s RNeasy mini spin columns were used for purification of the amplified cRNA samples. cRNA were quantified using NanoDrop Spectrophotometer version 3.2.1. 600 ng of cyanine 3-labelled, linearly amplified cRNA were used for hybridization. Hybridization was performed in a hybridization oven at 65°C for 17 hr at 10 rpm in SurePrint G3 Rat GE microarray slides (8 × 60K G4853A, Agilent Technologies, CA) containing 60,000 oligonucleotide probes. Hybridised microarray slides were then washed according to manufacturer’s instructions and scanned using an Agilent Microarray Scanner incorporating the Agilent Feature Extraction Software (Agilent Technologies). The microarray data is available from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the series accession number GSE67836.
Quantification files derived from the Agilent Feature Extraction Software were analyzed using the AgiND package (http://tagc.univmrs.fr/AgiND). We also used the AgiND R package for data quality control and normalization. Quantile methods and a background correction were used for data normalization. Gene expression data was then imported into XLStat (Addinsoft®) for filtering and statistical analyses. Because the primary objective was identifying genes suitable for being subjected to further bioinformatics analysis, all gene names containing the words «expressed sequence», «RIKEN», « PREDICTED», «cDNA sequence», or “similar to” were filtered out. Statistical analysis between groups was performed by unpaired unequal variance Welch’s t-test. Genes were considered to be statistically significant at p≤0.05. The false discovery rate (FDR), used to correct for errors in multiple testing [27,28], was estimated for each of these tests, using a single type I error cutoff (α = 0.05), as described previously [29-31].
Data from other microarray studies: Gene expression profiles in samples from the frontal cortex following simulated stress [32], depression [33], alcohol dependence [33], stroke [35], and Alzheimer’s disease [36] were collected directly from the original authors or downloaded from Gene Expression Omnibus (GEO) Datasets. All studies used adult rats while the Alzheimer’s disease model used transgenic mice.
Finding overlapping genes across pathobiological models: To test the possibility that gene expression profiles associated with brain trauma share some common patterns with other pathobiological models, we performed an overlap analysis of genes, comparing the present datasets with those of rodent models of stress, depression, alcohol dependence, stroke, and Alzheimer’s disease. We are aware that comparing across experimental platforms and analytical methods may introduce some variability. To counteract these caveats, we focused on a list of significant genes (at p≤0.05) across reported datasets rather than magnitudes of expression changes. Data sets from selected mentioned microarray studies were imported into Excel. To identify overlap genes a Venn diagram was generated based on Gene Symbol as primary field. Statistical analysis was performed using a binomial test [37].
Functional analysis of significant genes: Significant genes were classified into functional related gene groups using the Web-based gene ontology tool from the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.7 (http://david.abcc.ncifcrf.gov) [37-39]. This analysis was performed by using the two Functional Annotation tools, the Chart and the Clustering tool. The Chart report lists annotation terms and their associated genes and the Clustering tool group redundant annotation terms in clusters. Only Biological Process (BP) related functions were considered and default software settings were applied. A cut-off value of Enrichment Score of clusters was set at 2. To reduce redundancy, only a single significant BP (p≤0.05) that appeared to be the most informative and characteristic is reported for each cluster. Genes for each direction of change (up or down) were uploaded and analyzed separately, except when the number of significant genes was relatively small because it precluded a powerful analysis for over-represented BP categories.
The functional interactions between significant genes were analyzed by Gene Multiple Association Network Integration Algorithm (GeneMANIA) webserver (http://www.genemania.org) [40], which is a rapid and accurate heuristic algorithm that builds a composite network by integrating multiple functional associations. The BP category was used to create the interaction network. A graph was then built with one node for each gene and a link between genes that represent the co-expression profile. “Hub genes” can be identified; those that interact with a large number of genes in the current network and are most likely the biological key drivers.
Quantitative RT-PCR validation: To further confirm the reliability of the microarray data, the messenger RNA (mRNA) levels of 9 genes that were up or down regulated were quantified with qRT-PCR. These genes were activity-regulated cytoskeleton-associated protein (Arc; NM_019361; (F) CCCAGTCTGTGGCTTTTGTCA, (R) GTGTCAGCCCCAGCTCAATC), C-C motif chemokine ligand 3 (Ccl3; NM_013025, (F)GCCTGCTGCTTCTCCTACAG, (R)TCTGCCGGTTTCTCTTAGTC), bone morphogenetic protein 7 (Bmp7; NM_001191856; (F)CCTGGGCTTACAGCTCTCC, (R)TGCCAACTTGGGGTTGAT), ciliary neurotrophic factor (Cntf; NM_013166; (F)GACCTGACTGCTCTTATGGAATCT, (R)GCCTGGAGGTTCTCTTGGA), bone morphogenetic protein 4 (Bmp4; NM_012827; (F)GCATCCGAGCTGAGAGACCCCA, (R) ATCCCATCAGGGACGGAGACCA), vascular cell adhesion molecule 1 (Vcam1; NM_012889; (F)CAAATGGAGTCTGAACCCAAA, (R)GGTTCTTTCGGAGCAACG), caveolin 1 (Cav1; NM_133651; (F)AACGACGACGTGGTCAAGA, (R)CACAGTGAAGGTGGTGAAGC), annexin A2 (Anxa2; NM_019905; (F)CCTTCGCCTACCAGAGGAG, (R)CAGACAAGGCCGACTTCATC), aldehyde dehydrogenase 1 family, member A2 (Aldh1a2; NM_053896; (F)TCTCATGGTGTCCTCTGCAA, (R)TCTGAGCATTTAAGGCGTTG). The qRT-PCR assays were conducted according to the manufacturer's instructions (Ab Applied, France) using a LightCycler 480 Instrument with software version 1.5.0 (Roche, Meylan, France). Each reaction was run in duplicate in a final volume of 25 µl containing 12.5 µl of SYBR Green 2 X Master Mix (Roche Diagnostics, Mannheim, Germany), 300 nmol/l of each primer, and 5 µl of cDNA. PCR programmes were carried out as follows: one cycle at 95°C for 5 min and 45 cycles of amplification (95°C for 10 s, 60°C for 30 s, and 72°C for 15 s). The crossing point (Ct) of each set of primers was determined and compared to the Ct of the peptidylprolyl isomerase A (PpiA; NM_017101.1) mRNA used as endogenous controls. Subsequently data was imported into Excel for log2 transformation and statistical analyses. Linear regression analysis was then performed between qRT-PCR and microarray data using XLStat (Addinsoft®).
Gene expression profile in lateral fluid percussion brain injury rat model
One month after lfp-TBI, 1425 genes were significantly expressed in the frontal cortex and identified in knowledge banks (including PubMed, DAVID, Protein ANalysis THrough Evolutionary Relationships). Among them, 782 were up-regulated and 643 down-regulated (Table 1). The top 10 genes that had been up or down regulated are shown in Table 2.
From the up-regulated genes list, 279 significant biological processes (BP, see Method) were identified. These BP terms were grouped into four major categories, including metabolic process, immune system, cellular process, and morphogenesis (Figure 1A). Enriched BP terms in each of these categories are reported in Table 1.
Among the down-regulated genes list, two BP terms in particular were identified which belong to the metabolic and cellular processes (Figure 1A, Table 1).
Table .1 Functional processes identified in rats’ frontal cortex 1 month post lateral fluid percussion trauma. Significantly overrepresented up-regulated (left) and down-regulated (right) biological processes are indicated (p<0.05 by DAVID Functional Annotation Clustering tool). Genes belonging to each biological process are reported. Note that some of the genes fit within multiple categories.
Lateral fluid percussion Injury
1425 genes |
Up-regulated genes
782 genes |
Down-regulated genes
643 genes |
Metabolic processes |
Metabolic processes |
GO:0018107~peptidyl-threonine phosphorylation
Prkca, Sbk1, Tgfbr2, Tnks, Mtor
GO:0043408~regulation of MAPKKK cascade
Fgfr2, Ptprc, Ptpn6, Cav1, Tnf, Card9, Mylk2, Igf2, Timp2, Pdcd4, Itgb1, Tp73, Ctnnb1, Cdc42, Adrb2, Myd88, Mdfic
GO:0045055~regulated secretory pathway
Cadps, Lat, Unc13d, Steap2, Pex5l, Rab27a
Immune system
GO:0006954~inflammatory response
Bmp2, Ccl3, C5ar1, Tnf, Pdpn, C4b, Ephx2, Serping1, C1s, Tp73, Stat3, Vcam1, Lat, Tnfrsf1a, Cyba, Tnfrsf1b, Sdc1, Unc13d, Ptges, Fcgr1a, Cnr2, Cfh, Cd14, Fn1
GO:0002253~activation of immune response
Ptpn6, Ptprc, Myd88, C5ar1, Lax1, C4b, Cfh, Unc93b1, Serping1, C1s
GO:0019882~antigen processing and presentation
Icam1, Psme1, Tap2, Fcgr1a, Unc93b1, Ifi30, Rt1-M1-4, Rt1-Dmb, Cd1d1, Rt1-Bb
GO:0007159~leukocyte adhesion
Vcam1, Icam1, Ptprc, Tnf, Itga4, Msn, Itgb1
GO:0001906~cell killing
Ptpn6, Unc13d, Fcgr1a, Cyss, Rab27a
Cellular process
GO:0010647~positive regulation of cell communication
Fgfr2, Twsg1, Cav1, Acvrl1, Tnf, Slc20a1, Timp2, Itgb1, Ctnnb1, Cdc42, Tnfrsf1a, Myd88, Ghr, Ptprc, Cflar, Bmp2, Card9, Skp2, Igf2, Mbd2,
Sectm1a, Adrb2, Gria1, Tmod2, Mtor, Wnt9a, Bmp7, Bmpr1a
Morphogenesis
GO:0001763~morphogenesis of a branching structure
Fgfr2, Wnt5a, Bmp2, Nrp1, Tgfbr2, Mmp14, Gzf1, Ctnnb1, Vdr, Adm, Timeless, Phb2, Bmp7
GO:0048771~tissue remodeling
Nox4, Ctsk, Cav1, Adrb2, Rac2, Stat5a, Axl, Acp5, Itga4, Mmp14, Ctnnb1
GO:0016044~membrane organization
Cav1, Stx7, Rtp1, Tgfbr2, Colec12, Abca1, Cadps, Mfn2, Adrb2, Unc13d, Cd36, Gria1, Fcgr1a, Copb1, Rab34, Necap2, Sh3kbp1, Pafah1b1, Steap2, Ehd2, Ghr, Epn2
|
GO:0009063~cellular amino acid catabolic process
Aadat, Tdo2, Arg2, Gls, Gstz1, Prodh2, Pah, Fah
Cellular process
GO:0007268~synaptic transmission
Nos1ap, Slc12a5, Grik5, Lin7b, Acp1, Pclo, Ptk2, Dynll1, Npy, Gls, Sptbn2, Cd24, Cort, Fgf2, Lphn1
|
Table 2. Top 10 of most up or down-regulated significant genes in the cortex frontal of lateral fluid percussion injury rats. Gene symbols, names, ID, LogRatio, P-value, and Go Biological process description are provided.
|
Gene symbol
|
Gene name
|
Gene ID
|
LogR
|
P-Value
|
GO Biological process description
|
|
Up-regulated gene
|
|
|
|
|
|
|
Igf2
|
insulin-like growth factor 2
|
NM_031511
|
1.73
|
0.00
|
GO:0010647~positive regulation of cell communication
|
|
RT1-Bb
|
RT1 class II, locus Bb
|
NM_001004084
|
1.44
|
0.01
|
GO:0019882~antigen processing and presentation
|
|
Ptges
|
prostaglandin E synthase
|
NM_021583
|
1.37
|
0.00
|
GO:0006954~inflammatory response
|
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic
|
NM_022936
|
1.02
|
0.01
|
GO:0006954~inflammatory response
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
NM_199093
|
1.01
|
0.01
|
GO:0006954~inflammatory response
|
|
Tp73
|
tumor protein p73
|
NM_001108696
|
0.90
|
0.00
|
GO:0006954~inflammatory response
|
|
Bmp7
|
bone morphogenetic protein 7
|
NM_001191856
|
0.89
|
0.01
|
GO:0010647~positive regulation of cell communication
|
|
Fn1
|
fibronectin 1
|
NM_019143
|
0.85
|
0.01
|
GO:0006954~inflammatory response
|
|
C4b
|
complement component 4B
|
NM_031504
|
0.85
|
0.00
|
GO:0006954~inflammatory response
|
|
C1s
|
complement component 1, s subcomponent
|
NM_138900
|
0.84
|
0.00
|
GO:0002253~activation of immune response
|
|
|
|
|
|
|
|
|
Down-regulated gene
|
|
|
|
|
|
|
Cort
|
cortistatin
|
NM_012835
|
-0.58
|
0.01
|
GO:0007268~synaptic transmission
|
|
Pah
|
phenylalanine hydroxylase
|
NM_012619
|
-0.43
|
0.01
|
GO:0009063~cellular amino acid catabolic process
|
|
Prodh2
|
proline dehydrogenase 2
|
NM_001038588
|
-0.35
|
0.01
|
GO:0009063~cellular amino acid catabolic process
|
|
Aadat
|
aminoadipate aminotransferase
|
NM_017193
|
-0.30
|
0.00
|
GO:0009063~cellular amino acid catabolic process
|
|
Fgf2
|
fibroblast growth factor 2
|
NM_019305
|
-0.28
|
0.01
|
GO:0007268~synaptic transmission
|
|
Fah
|
fumarylacetoacetate hydrolase
|
NM_017181
|
-0.27
|
0.01
|
GO:0009063~cellular amino acid catabolic process
|
|
Cd24
|
CD24 molecule
|
NM_012752
|
-0.23
|
0.01
|
GO:0007268~synaptic transmission
|
|
Npy
|
neuropeptide Y
|
NM_012614
|
-0.23
|
0.00
|
GO:0007268~synaptic transmission
|
|
Lin7b
|
lin-7 homolog b
|
NM_021758
|
-0.22
|
0.01
|
GO:0007268~synaptic transmission
|
|
Pclo
|
piccolo
|
NM_020098
|
-0.22
|
0.00
|
GO:0007268~synaptic transmission
|
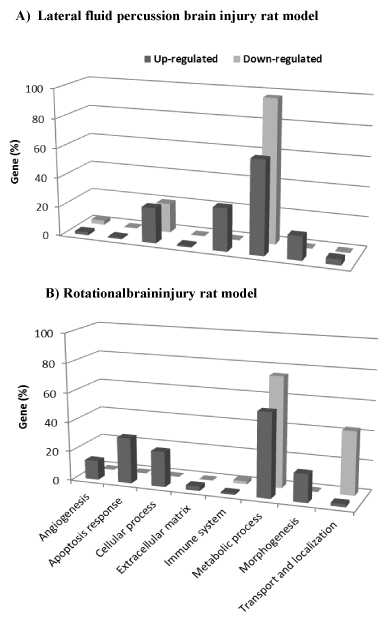
Figure 1. Functional gene categories of significant genes related to lateral fluid percussion (A) and rotational injuries (B). Eight functional categories of genes were identified using DAVID Functional Annotation Chart tool. The histograms represent the percentage of significant genes belonging to the functional category.
Gene expression profile in rotational brain injury rat model
Rotational injury induced expression changes for 965 genes identified in the knowledge banks; 457 were up-regulated and 508 were down-regulated (Table 3). The top 10 genes that had been up or down regulated are shown in Table 4.
Table 3. Functional processes identified in rats’ frontal cortex 1 month post rotational trauma. Significant overrepresented up-regulated (left) and down-regulated (right) biological processes are indicated (p<0.05 by DAVID Functional Annotation Clustering tool). Genes belonging to each biological process are reported. Note that some of the genes fit within multiple categories.
| 965 genes |
Up-regulated genes
457 genes |
Down-regulated genes
508 genes |
Apoptosis response
GO:0042981~regulation of apoptosis
Siva1, Xrcc5, Nfkbia, Prdx1, Ptma, Ada, Timp1, Aldh1a1, Bak1, Apoe, Agt, Casp7, Psenen, Bcl6, Inpp5d, Fn1, Egfr, Txnip, Sgk1, Cd3g, Cryab, Rxra, Sf1, Csda, Plekhf1, Cdh13, Msx1, Cntf, Nupr1, Dusp1, Igf2r, Ube2m, Bmp7, Abl1, Eif2ak3 |
Metabolic process
GO:0033674~positive regulation of kinase activity
Irak1, Prkcz, Taok2, Madd, Hgf, Adora1, Cdc25b, Akt1, Prkar2b, Lrp1, Mapk8ip3, Cd4, Jak2, Traf7, Crk |
Morphogenesis
GO:0048729~tissue morphogenesis
Egfr, Bmp4, Rxra, Ptk7, Snai1, Aldh1a1, Tnnt2, Aldh1a2, Adm, Agt, Col1a2, Gatad2a, Bmp7, Tbx18, Twist1, Alx1
GO:0016044~membrane organization
Arc, Hsp90aa1, Cltb, Cubn, Hip1r, Snx17, Lmna, Colec12, Col5a1, Zdhhc15, Bak1, March2, Dab2, Rabep2, Apoe, Rab34, Snap23 |
Transport and Localization
GO:0046907~intracellular transport
Pabpn1, Arl1, Prkcz, Xpo6, Ap1b1, Ywhab, Ap3s1, Bnip3, Ipo9, Nxf1, Pola2, Vti1a, Atxn1, Akt1, Cog3, Ap1s1, Gphn, Zfp280b, Scyl1, Tap1, Exoc4, Jak2, Mybbp1a, Tob1 |
Angiogenesis
GO:0001944~vasculature development
Reck, Bmp4, Anpep, Elk3, Col5a1, Anxa2, Bak1, Cdh13, Aldh1a2, Apoe, Ctgf, Agt, Col1a2, Gatad2a |
|
Extracellular matrix
GO:0030198~extracellular matrix organization
Reck, Lgals3, Ccdc80, Vtn, Serpinh1, Col5a1, Anxa2, Emilin1, Smoc2, Kazald1, Agt, Itga8, Col1a2, Adamts2, Fn1 |
|
|
|
Table 4. Top 10 of most up or down-regulated significant genes in the cortex frontal of rotational trauma rats. Gene symbols, names, ID, LogRatio, P-value, and Go Biological process description are provided.
Gene symbol |
Gene name |
Gene ID |
LogR |
p-value |
GO Biological process description |
Up-regulated gene |
|
|
|
|
|
Bmp7 |
bone morphogenetic protein 7 |
NM_001191856 |
0.91 |
0.01 |
GO:0048729~tissue morphogenesis |
Tbx18 |
T-box18 |
NM_001108173 |
0.92 |
0.01 |
GO:0048729~tissue morphogenesis |
Msx1 |
msh homeobox 1 |
NM_031059 |
0.96 |
0.00 |
GO:0042981~regulation of apoptosis |
Tnnt2 |
troponin T type 2 |
NM_012676 |
0.97 |
0.01 |
GO:0048729~tissue morphogenesis |
Txnip |
thioredoxin interacting protein |
NM_001008767 |
0.98 |
0.01 |
GO:0042981~regulation of apoptosis |
Alx1 |
ALX homeobox 1 |
NM_012921 |
1.02 |
0.00 |
GO:0048729~tissue morphogenesis |
Col1a2 |
collagen, type I, alpha 2 |
NM_053356 |
1.20 |
0.01 |
GO:0048729~tissue morphogenesis |
Nupr1 |
nuclear protein, transcriptional regulator, 1 |
NM_053611 |
1.23 |
0.01 |
GO:0042981~regulation of apoptosis |
Aldh1a2 |
aldehyde dehydrogenase 1 family, member A2 |
NM_053896 |
1.36 |
0.01 |
GO:0048729~tissue morphogenesis |
Cd3g |
CD3 molecule, gamma |
NM_001077646 |
1.41 |
0.00 |
GO:0042981~regulation of apoptosis |
Down-regulated gene |
|
|
|
|
|
Cd4 |
Cd4 molecule |
NM_012705 |
-0.75 |
0.00 |
GO:0033674~positive regulation of kinase activity |
Hgf |
hepatocyte growth factor |
NM_017017 |
-0.58 |
0.01 |
GO:0033674~positive regulation of kinase activity |
Cog3 |
component of oligomeric golgi complex 3 |
NM_001012157 |
-0.51 |
0.01 |
GO:0046907~intracellular transport |
Tob1 |
transducer of ErbB-2.1 |
NM_133317 |
-0.46 |
0.00 |
GO:0046907~intracellular transport |
Tap1 |
transporter 1, ATP-binding cassette, sub-family B |
NM_032055 |
-0.45 |
0.01 |
GO:0046907~intracellular transport |
Ipo9 |
importin 9 |
NM_001107180 |
-0.35 |
0.00 |
GO:0046907~intracellular transport |
Taok2 |
TAO kinase 2 |
NM_022702 |
-0.33 |
0.01 |
GO:0033674~positive regulation of kinase activity |
Adora1 |
adenosine A1 receptor |
NM_017155 |
-0.29 |
0.01 |
GO:0033674~positive regulation of kinase activity |
Irak1 |
interleukin-1 receptor-associated kinase 1 |
NM_001127555 |
-0.25 |
0.00 |
GO:0033674~positive regulation of kinase activity |
Xpo6 |
exportin 6 |
NM_001011935 |
-0.25 |
0.01 |
GO:0046907~intracellular transport |
The DAVID Functional Annotation Clustering tool identified 104 significant BP terms in the list of up-regulated genes. Most of these genes were associated with apoptosis, morphogenesis, angiogenesis and extracellular matrix organization categories (Figure 1B). The BP enriched terms identified are reported in Table 3.
Down-regulated genes belong to the metabolic process and transport and localization categories (Figure. 1B). Two BP terms were significantly overrepresented as shown in Table 3.
Comparison across trauma models
To determine whether the two trauma models showed similarities in expression changes, a Venn diagram was generated from the significant genes in lfp-TBI and rot-TBI frontal cortex at 1 month post-trauma, which facilitated identification of 79 common genes (p<0.05; binomial test). Note that for a functional profile, up and down-regulated genes were analyzed together because of the limited number of overlapping genes. Only cell adhesion which included seven genes (protocadherin alpha 2, Pcdha2; hes family bHLH transcription factor 5, Hes5; limbic system-associated membrane protein, Lsamp; tweety family member 1, Ttyh1; receptor tyrosine kinase-like orphan receptor 2, Ror2; protocadherin gamma subfamily A2, Pcdhga2; and fibronectin1, Fn1 genes) was found to be over-represented.
Comparison to other pathobiological models
Complete microarray datasets are available for rat brain studies of depression (GSE20388) [33] and alcohol dependence (GSE4494) [34]. Although three studies examined gene expression changes in rat cortex following stress [32], stroke [33], and Alzheimer’s disease [34] only lists of significant genes, rather than complete data sets, are available for these studies. Data are reported in Figure 2 and Table 5.
Following lfp-TBI, stroke represented the pathology showing the most overlap of genes, with 25% of homology, i.e., of the 1425 genes identified 1 month post lfp-TBI, 357 were identical to those reported in cortex following stroke (p<0.05; binomial test). In particular, these common genes were involved in biological activities related to immune/inflammation response and matrix organization as well as being subjected to network construction. GeneMANIA was used to investigate how these genes interact with each other and to identify hub genes (see Method). Among genes belonging to the leukocyte migration category, C-C motif chemokine ligand 3 (Ccl3) interacted the most (node degree of 4). Serpin peptidase inhibitor, clade G, member 1 gene (Serping1) and protein tyrosine phosphatase non-receptor type 6 (Ptpn6) were predicted by GeneMANIA as hub genes in the immune response network category (both having the node degree of 14). Interestingly, the Ptpn6 gene was also classified in the response to wounding category and identified as the gene with the highest interaction (node degree of 17). In both networks, Ptpn6 was linked to genes such as complement component 1S (C1s) and complement component 4B (C4b). Note that C1s was also identified as a hub gene in the response to the wounding network category (node degree of 17). For the skeletal system category, matrix metallopeptidase 14 (Mmp14) was considered as the hub gene (node degree of 9). The collagen, type I, alpha 1 (Col1a1) gene interacted the most in the extracellular matrix network category (node degree of 5). Genes belonging to the membrane organization category did not interact with each other.
Nineteen percent of the genes that were significantly expressed post focal trauma were similar to those identified in the genetic rat model of depression (p<0.05; binomial test) (Figure 2 and Table 5) and involved in intracellular transport and cell adhesion. Network analysis showed that among genes belonging to the intracellular transport, syntaxin 7 (Stx7) interacted the most (node degree of 5). Podoplanin, node degree of 5 (Pdpn) was the gene that interacted the most in cell adhesion and it interacted with vascular cell adhesion molecule-1 gene (Vcam1).
Compared with the other models studied (stress, alcohol dependence, and Alzheimer’s disease), less than 10% of gene homology was identified (p>0.05; binomial test) (Figure 2 and Table 5).
Following rot-TBI, similar analyses as for lfp-TBI were carried out. Stroke and depression were the two pathologies showing the highest overlap of significantly expressed genes, with 24% and 16% homology, respectively (Figure 2 and Table 5).
There were 232 common genes between rot-TBI and stroke (p<0.05; binomial test) and they were associated with extracellular matrix organization and regulation of cell growth. Network analysis showed that among genes belonging to extracellular matrix organization, collagen, type I, alpha 2 (Col1a2) and serpin peptidase inhibitor H1 genes (Serpinh1) interacted the most (node degree of 5). The gene interacting most with regulation of cell growth was the disabled homolog 2, node degree of 3 (Dab2) gene.
There were 157 overlapping genes between rot-TBI and depression which are related to the intracellular signalling cascade (p<0.05; binomial test) (Figure 2 and Table 5). The glutamate receptor, metabotropic 7 (Grm7) gene was identified as a hub gene (node degree of 4).
Less than 10% of the other pathobiological models (stress, alcohol dependence, and Alzheimer’s disease) used in the present study had common genes that were significantly expressed (p>0.05; binomial test) (Figure 2 and Table 5).
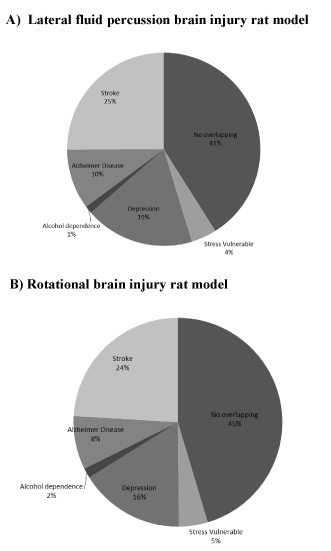
Figure 2. Pie charts indicate percentage of genes assigned to each pathobiological model for the two trauma models (A, lateral fluid percussion and B, rotational injuries).
Table 5. Gene number and gene ontology analysis of overlapping genes between the lateral fluid percussion or rotational injuries and others pathobiological models. Genes belonging to each biological process are reported. Underlined gene symbols are hub genes identified within GeneMania networks.
|
Lateral fluid percussion injury |
Rotational injury |
Others pathobiologies |
|
|
Stress |
# 61 genes
GO:0042127~regulation of cell proliferation (Fgfr2, Cyba, Adrb2, Anp32b, Phb2, Gata4, Bmp7) |
#43 genes
GO:0048514~blood vessel morphogenesis (Bmp4, Ctgf, Apoe, Anxa2) |
Depression |
# 261 genes
GO:0046907~intracellular transport (Dync1i1, Stx8, Stx7, Arntl, Flna, Bcap31, Cog3, Cdc42, Npc2, Chic2, Mlx, Pafah1b1, Sec23b, Sec61a1, Rab27a, Ssr3)
GO:0007155~cell adhesion (Col18a1, Aebp1, Ppard, Nrp1, Pdpn, Lef1, Itgb1, Vcam1, Cdh8, Cdh22, Azgp1, Cdc42, Lamb3, Gp1bb, Itgb7, Ttyh1, Aoc3, Fn1)
|
# 157 genes
GO:0007242~intracellular signaling cascade (Rab3b, Fgfr3, Stk38, Gnai1, Nfkbia, Adora1, Plcl2, Apoe, Ctgf, Rhobtb2, Gng3, Dnaja3, Egfr, Arl1, Madd, Rxra, Rab4b, Arl6, Trim23, Cdh13, Arf2, Rras2, Grm7, Rab5a, Nkiras1) |
Alcohol dependence |
# 18 genes
No cluster with Enrichment Score ≥2 |
# 15 genes
GO:0031589~cell-substrate adhesion (Agt, Vtn, Fn1) |
Stroke |
#357 genes
GO:0050900~leukocyte migration (Prkca, Icam1, Ccl3, C5ar1, Il16, Cklf, Cd24, Msn)
GO:0006955~immune response (Gpr183, Ccl3, Acp5, C1s, Sp110, Cd1d1, Rt1-Bb, Tnfrsf1b, Myd88, Tap2, Fcgr1a, Cfh, Bcl3, Cd24, Rab27a, Ptprc, Icam1, Ptpn6, Card9, C4b, Serping1, Cyba, Unc13d, Cxcl13, Lilrb3, Cd14)
GO:0009611~response to wounding (Gfap, Ccl3, Acvrl1, Nrp1, C1s, Dcn, Pcsk1, Tnfrsf1a, Tnfrsf1b, Gsn, Il10rb, Ptges, Gp1bb, Fcgr1a, Cnr2, Cfh, Cd24, Entpd1, Fgf2, Rab27a, Fn1, Ptpn6, C5ar1, Pdpn, C4b, Tgfbr2, Serping1, Sparc, Stat3, Cyba, Sdc1, Unc13d, Adm, Ctsb, Cd14)
GO:0001501~skeletal system development (Prkca, Wnt5a, Twsg1, Lgals3, Mmp9, Hexb, Tgfbr2, Cytl1, Acp5, Igf2, Sparc, Mmp14, Ctsk, Col1a2, Col1a1, Gpnmb, Fn1, Ghr)
GO:0016044~membrane organization (Arc, Tgfbr2, Abca1, Cadps, Asgr2, Adrb2, Unc13d, Cd36, Ulk1, Fcgr1a, Necap2, Cd24, Steap2, Ehd2, Ghr)
GO:0030198~extracellular matrix organization (Col18a1, Lgals3, Mmp9, Col1a2, Olfml2a, Bcl3, Dcn, Col1a1, Fn1, Emilin1)
|
# 232 genes
GO:0030198~extracellular matrix organization (Lgals3, Agt, Kazald1, Col1a2, Serpinh1, Fn1, Anxa2, Emilin1)
GO:0001558~regulation of cell growth (Nov, Acvr1b, Sgk1, Dab2, Agt, Kazald1, Igfbp2) |
Alzheimer disease |
# 144 genes
GO:0009611~response to wounding (Ptpn6, Ccl3, Nrp1, C4b, Tgfbr2, Igf2, Stat3, Tnfrsf1a, Cyba, Tnfrsf1b, Cnr2, Entpd1, Cd14)
GO:0006470~protein amino acid dephosphorylation (Ppp6c, Ptpn6, Ptprc, Dusp1, Ppm1b, Mtmr7)
GO:0008219~cell death (Ptpn6, Dffa, Igf2, Pdcd4, Pdcd1, Dapk1, Tnfrsf1a, Clptm1l, Tnfrsf1b, Ripk3, Ctsd, Rnf34, Cd14)
GO:0050727~regulation of inflammatory response (Zfp36, Tnfrsf1a, Tnfrsf1b, Osmr, Igf2)
|
# 80 genes
GO:0001501~skeletal system development (Sgpl1, Chrdl1, Msx1, Lgals3, Ctgf, Foxc2, Igf2)
GO:0001525~angiogenesis (Ctgf, Foxc2, Hs6st1, Elk3) |
Quantitative RT-PCR validation
We validated the microarray results with qRT-PCR using primers and probes for a subset of genes (see Figure. 3), which showed that gene expression changes measured with qRT-PCR correlate strongly with changes found through microarray analysis (R2=0.67; F(1,8)=14.32; P<0.007).
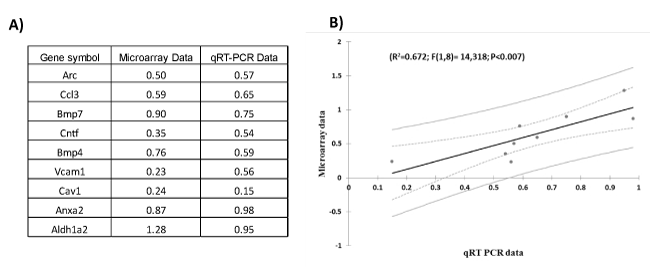
Figure 3. Quantitative real-time PCR validation of the microarray data for genes related to TBI. A) Table indicates the fold-change (in Log2 scale) of genes measured by microarray and qRT-PCR experiments. B) Regression plot between qRT PCR and microarray data for genes in frontal cortex of lateral fluid percussion injury and rotational brain trauma.
The present study was designed to identify the molecular signature in traumatized brains and to compare them with brains that had been subjected to stress, depression, alcohol dependence, stroke, and Alzheimer’s disease. Data showed that the two trauma models, lfp-TBI and rot-TBI, had genes in common with depression and stroke models.
Changes in expression of hundreds of genes following brain trauma were revealed in the present study. Several other studies reported in literature used microarray to examine rodent brains post TBI. However, the majority of these studies focused on genes and pathway changes in the brain tissue a few hours post injury [16,17,19,21,42-45]. Only a few sets of microarray data are available for longer post-injury periods. Von Gertten et al. showed time dependence in genomic responses between 1 to 4 days following cerebral cortical contusion in rat cortex [20]. In mouse, gene expression changes were studied up to 14 days after trauma by Kobori et al. [46] in the cortex and up to 40 days after trauma by Tweedie et al. [47] in the hippocampus. To the best of our knowledge, this is the first study to examine gene expression profiles following traumatic brain injury at a post-trauma time as extended as 1 month in the rat frontal cortex. Hence, our data indicates that transcriptional alterations still occur 1 month post injury and, in addition, in brain regions not located close to the lesion. Gene changes in the TBI penumbra have been revealed by Darkazalli et al. [48] in a rodent model of controlled cortical impact. Functional analysis revealed that this TBI regulated a large number of cortical genes belonging to pathways involved in metabolism and immune cell notably. One month following lfp-TBI, our microarray data in frontal cortex indicated expression changes of genes related to immune system. In particular, we showed an up-regulation of C-C motif chemokine ligand 3 (Ccl3) and tumor necrosis factor (Tnf) genes, i.e., genes that Kim et al. [48] reported to be up-regulated in macrophages isolated from the brain hemisphere of mouse following TBI . Innate immune response activation was also reported after sports-related concussion within 6 h in human peripheral blood mononuclear cells [49]. Following rot-TBI, our data revealed an up-regulation of biological processes related to apoptosis, comforting data of Lončarević-Vasiljković et al. [50]. In a recent study, processes related to cell death were identified in blood samples collected from patients with TBI within the first 24 h of injury [51].
Overall, lfp-TBI and rot-TBI generated a specific molecular profile. Such results might be explained by differences in initiating triggers of trauma but also injury severity or that the recovery trajectory differed between the two models. When related biological processes were considered no similarity was observed, except the one associated with morphogenesis. Thus, the two trauma models have mechanisms belonging to tissue remodelling and membrane organization in common, i.e., processes that might be associated with tissue protection and repair. Of particular interest is the recent study of Cho et al. [52]. Examining gene expression changes from peripheral blood of human TBI patients, they suggest that gene activity promotes TBI recovery. These processes are initiated as soon as 48h after trauma. Genes related to neuronal recovery were also identified in the peripheral blood of military personnel with a history of blast-TBI during deployment [53].
When cognitive deficits post trauma are considered, it has been observed in rodents that they persist for up to 4 weeks or longer following lateral fluid percussion injury [11]. Following rotational head injury, Stemper et al. indicated that rats had cognitive deficits when tested in an elevated plus maze [54]. In the present study, the behavior of rats was not tested, hence clear conclusion could not be made. Nonetheless, the spontaneous reparative capabilities identified through gene functional pathways, if they exist, would not be sufficient to annihilate behavioral deficits.
2021 Copyright OAT. All rights reserv
Depressive disorders are the most frequent neuropsychiatric complication of TBI [55,56]. Depression is closely associated with selective structural changes, altered cellular resilience, and neuronal atrophy [57]. In the frontal cortex in particular, alterations in protein synthesis, neuromediator regulations, and ion equilibrium have been reported [58]. The present study showed that TBI, no matter what the trauma model was, and depression share similar gene expression changes, indicating a common molecular support between these states. Pharmacologically, this has a major impact. To date, the treatment of depression secondary to TBI is often approached with serotonin reuptake inhibitors [59]. The present study aimed to identify more specific therapeutic targets post TBI. To this end, the genes whose expression were altered in both TBI and depression rat models were assumed to be potential therapeutic targets for depression in TBI. The focus among genes involved in both TBI and depression was on hub genes. Indeed, hub genes are expected to play an important role in biology in regard to their frequent interaction with others genes. Hence, these genes were, notably, syntaxin 7 (Stx7), which is known as an essential mediator of exocytosis of synaptic vesicles in neurons [60], podoplanin (Pdpn), which plays a role in vascular development and homeostasis [61], vascular cell adhesion molecule 1 (Vcam1), which is critical for subventricular zone maintenance and function [62], and glutamate receptor, metabotropic 7 (Grm7), which is involved in the neurotransmission of glutamate. Potentiation of one of these genes or a combination of some of them can be expected to be of therapeutic benefit. For instance, the use of Pdpn and Grm7 would provide a permissive environment for cellular regeneration post brain injury.
Stroke such as cerebral ischemia used in the present study is considered as one of a number of secondary injury mechanism of TBI [63]. Stroke and TBI have been shown to cause similar brain alterations, such as inflammation, apoptotic cell death, and white matter damaged notably. Treatment of stroke in chronic TBI relies on the use of aspecific agents, such as beta blockers or agonists and drugs that target the catecholaminergic system in particular [55,11]. The present study helps to identify genes which might be considered as novel therapeutic targets for the treatment of stroke in TBI. For instance, focus should be on the C-C motif chemokine ligand 3 (Ccl3), serpin peptidase inhibitor, clade G, member 1 (Serping1), complement component 1S (C1s) and protein tyrosine phosphatase non-receptor type 6 (Ptpn6) genes, which are common genes involved in immune/inflammation processes. An example of this is a recent study in a mouse TBI model 1 day post-injury which showed that Ptpn6 is known to be a signalling molecule acting as a regulator for a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Here, Ptpn6 was associated with the complement proteins (C1s and C4b genes) known to play a significant role in the regulation of T-cell responses [64]. Other genes might be targeted such as matrix metallopeptidase 14 (Mmp14), collagen, type I, alpha 1 and 2 (Col1a1 and 2) and disabled homolog 2 (Dab2), which were identified in the networks related to brain tissue remodelling. Matrix metallopeptidase such as Mmp14 are known to be involved in diverse physiological processes including tissue growth and regeneration, angiogenesis, apoptosis, and nerve growth [65]. Col1a1 and 2 play a critical role in the pathology of intracerebral haemorrhages. Dab2 plays an important role in cell signalling and migration [66]. Overexpression of such genes involved in functions related to brain tissue maintenance has the potential to have significant clinical potential.
Stress, alcohol dependence, and Alzheimer’s disease in rodent models did not significantly share genes with traumatized rats (neither lfp- or rot-TBI), suggesting that the relationship between these pathobiologies and TBI are not molecular in nature. However, we should emphasise that only one post-trauma time of 1 month was studied while neurological disorders, especially such as addiction and Alzheimer’s disease, would appear years after trauma. Alternatively, one may speculate that brain alterations generated by a trauma lead to tissue vulnerability in favor of these secondary medical complications. Yet, the link between these sequelae and TBI still remains to be elucidated.
Altogether, the data of the current study suggest that alterations in the molecular signatures are present 1 month post TBI in brain regions not directly affected by the injury such as frontal lobe. Gene expression profiles following TBI shared similar changes as those during depression and after stroke, indicating a common molecular support between these states. The biological implications of theses overlapping genes still remain to be elucidated. It should be emphasized that the data analyses used in the present study do not provide proof of gene involvement, but they do provide suggestions that support in directing further research. In particular, it will be necessary to test trauma rats to identify cognitive deficits and behavioral troubles related to depressive disorders and stroke. Thus rats will be submitted to a collection of tests and various measures will be extracted, such as the consumption of sweets to assess the hedonic state of the rat, appetite and food intake, performances in a forced swim test and locomotor activities in an open-field, and motor coordination. The time at which rats would be tested need to be determined.
This research was supported by the French Ministry for Education and Research funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Author contributions are as follow: conceived and designed the experiments: VP, CC; data analysis: VP; reagents/materials contribution: MO, JD, NF; paper writing: VP; manuscript supervision: MR, JD, BAL. We are grateful to Dr. E.M. Blalock for microarray analysis advice and to Dr. C. Benatti, Dr. K. Bosser, and Dr. M.W. Kimpel for providing microarrays data sheets. There is no conflict of interest.
- Gardner RC, Burke JF, Nettiksimmons J, Kaup A, Barnes DE5, et al. (2014) Dementia risk after traumatic brain injury vs nonbrain trauma: the role of age and severity. JAMA Neurol 71: 1490-1497. [Crossref]
- Katz DI, Cohen SI, Alexander MP (2015) Mild traumatic brain injury. Handb Clin Neurol 127: 131-156. [Crossref]
- Marshall KR, Holland SL, Meyer KS, Martin EM, Wilmore M, et al. (2012) Mild traumatic brain injury screening, diagnosis, and treatment. Mil Med 177: 67-75. [Crossref]
- Niemeier JP, Grafton LM, Chilakamarri T (2015) Treating persons with traumatic brain injury: history and updates. N C Med J 76: 105-110. [Crossref]
- Chauhan NB1 (2014) Chronic neurodegenerative consequences of traumatic brain injury. Restor Neurol Neurosci 32: 337-365. [Crossref]
- Masel BE, DeWitt DS (2010) Traumatic brain injury: a disease process, not an event. J Neurotrauma 27: 1529-1540. [Crossref]
- Johnson VE, Stewart W, Smith DH (2012) Widespread t and amyloid-ß pathology many years after a single traumatic brain injury in humans. Brain Pathol Zurich Switz 22: 142–149. [Crossref]
- Uryu K, Laurer H, McIntosh T, Praticò D, Martinez D, et al. (2002) Repetitive mild brain trauma accelerates Abeta deposition, lipid peroxidation, and cognitive impairment in a transgenic mouse model of Alzheimer amyloidosis. J Neurosci 22: 446–454. [Crossref]
- Lamprecht MR, Elkin BS1,, Kesavabhotla K, Crary JF, Hammers JL, et al. (2016) Strong Correlation of Genome-Wide Expression after Traumatic Brain Injury In Vitro and In Vivo Implicates a Role for SORLA. J Neurotrauma. [Crossref]
- Burke JF, Stulc JL, Skolarus LE, Sears ED, Zahuranec DB, et al. (2013) Traumatic brain injury may be an independent risk factor for stroke. Neurology 81: 33-39. [Crossref]
- Osier ND, Dixon CE (2015) Catecholaminergic based therapies for functional recovery after TBI. Brain Res 1640: 15-35. [Crossref]
- Carré E, Cantais E, Darbin O, Terrier JP, Lonjon M, et al. (2004) Technical aspects of an impact acceleration traumatic brain injury rat model with potential suitability for both microdialysis and PtiO2 monitoring. J Neurosci Methods 140: 23–28. [Crossref]
- Kabadi SV, Hilton GD, Stoica BA, Zapple DN, Faden AI (2010) Fluid-percussion-induced traumatic brain injury model in rats. Nat Protoc 5: 1552-1563. [Crossref]
- Xiong Y, Mahmood A, Chopp M (2013) Animal models of traumatic brain injury. Nat Rev Neurosci 14: 128-142. [Crossref]
- Risling M, Davidsson J (2012) Experimental animal models for studies on the mechanisms of blast-induced neurotrauma. Front Neurol 3: 30. [Crossref]
- Colak T, Cine N, Bamac B, Kurtas O, Ozbek A, et al. (2012) Microarray-based gene expression analysis of an animal model for closed head injury. Injury 43: 1264-1270. [Crossref]
- Risling M, Plantman S, Angeria M, Rostami E, Bellander BM, et al. (2011) Mechanisms of blast induced brain injuries, experimental studies in rats. Neuroimage 54 Suppl 1: S89-97. [Crossref]
- Sabir M, Gaudreault PO, Freyburger M, Massart R, Blanchet-Cohen A, et al. (2015) Impact of traumatic brain injury on sleep structure, electrocorticographic activity and transcriptome in mice. Brain Behav Immun 47: 118-130. [Crossref]
- Shojo H, Kaneko Y, Mabuchi T, Kibayashi K, Adachi N, et al. (2010) Genetic and histologic evidence implicates role of inflammation in traumatic brain injury-induced apoptosis in the rat cerebral cortex following moderate fluid percussion injury. Neuroscience 171: 1273–1282. [Crossref]
- von Gertten C, Flores Morales A, Holmin S, Mathiesen T, Nordqvist AC (2005) Genomic responses in rat cerebral cortex after traumatic brain injury. BMC Neurosci 6: 69. [Crossref]
- White TE, Surles-Zeigler MC, Ford GD, Gates AS, Davids B, et al. (2016) Bilateral gene interaction hierarchy analysis of the cell death gene response emphasizes the significance of cell cycle genes following unilateral traumatic brain injury. BMC Genomics 17: 130. [Crossref]
- Dalley JW, Cardinal RN, Robbins TW (2004) Prefrontal executive and cognitive functions in rodents: neural and neurochemical substrates. Neurosci Biobehav Rev 28: 771-784. [Crossref]
- McAllister TW, Sparling MB, Flashman LA, Guerin SJ, Mamourian AC, et al. (2001) Differential working memory load effects after mild traumatic brain injury. Neuroimage 14: 1004-1012. [Crossref]
- Turner GR, McIntosh AR, Levine B (2011) Prefrontal Compensatory Engagement in TBI is due to Altered Functional Engagement of Existing Networks and not Functional Reorganization. Front Syst Neurosci 5: 9-12. [Crossref]
- Davidsson J, Risling M (2011) A new model to produce sagittal plane rotational induced diffuse axonal injuries. Front Neurol 2: 41. [Crossref]
- Paxinos G, Watson C (1998) The rat brain in stereotaxic coordinates. (4th edtn). Academic Press, San Diego
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279-284. [Crossref]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinforma Oxf Engl 19: 368–375. [Crossref]
- Paban V, Chambon C, Farioli F, Alescio-Lautier B (2011) Gene regulation in the rat prefrontal cortex after learning with or without cholinergic insult. Neurobiol Learn Mem 95: 441-452. [Crossref]
- Paban V, Farioli F, Romier B, Chambon C, Alescio-Lautier B (2010) Gene expression profile in rat hippocampus with and without memory deficit. Neurobiol Learn Mem 94: 42-56. [Crossref]
- Benatti C, Valensisi C, Blom JM, Alboni S, Montanari C, et al. (2012) Transcriptional profiles underlying vulnerability and resilience in rats exposed to an acute unavoidable stress. J Neurosci Res 90: 2103–2115. [Crossref]
- Blaveri E, Kelly F, Mallei A, Harris K, Taylor A, et al. (2010) Expression profiling of a genetic animal model of depression reveals novel molecular pathways underlying depressive-like behaviours. PloS One 5: e12596. [Crossref]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, et al. (2007) Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol Fayettev N 41: 95–132.
- Ramos-Cejudo J, Gutiérrez-Fernández M, Rodríguez-Frutos B, Expósito Alcaide M, Sánchez-Cabo F, et al. (2012) Spatial and temporal gene expression differences in core and periinfarct areas in experimental stroke: a microarray analysis. PloS One 7: e52121. [Crossref]
- Wirz KT, Bossers K, Stargardt A, Kamphuis W, Swaab DF, et al. (2013) Cortical beta amyloid protein triggers an immune response, but no synaptic changes in the APPswe/PS1dE9 Alzheimer’s disease mouse model. Neurobiol Aging 34: 1328–1342. [Crossref]
- Blalock EM, Grondin R, Chen KC, Thibault O, Thibault V, et al. (2010) Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J Neurosci 30: 6058–6071. [Crossref]
- Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [Crossref]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44-57. [Crossref]
- Mehta JP, Rani S (2011) Software and tools for microarray data analysis. Methods Mol Biol 784: 41-53. [Crossref]
- Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q (2014) GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res 3: 153. [Crossref]
- Dash PK, Kobori N, Moore AN (2004) A molecular description of brain trauma pathophysiology using microarray technology: an overview. Neurochem Res 29: 1275–1286. [Crossref]
- Long Y, Zou L, Liu H, Lu H, Yuan X (2003) Altered expression of randomly selected genes in mouse hippocampus after traumatic brain injury. J Neurosci Res 71: 710–720. [Crossref]
- Natale JE, Ahmed F, Cernak I, Stoica B, Faden AI (2003) Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J Neurotrauma 20: 907–927. [Crossref]
- Zhang XY, Gu CG, Gu JW, Zhang JH, Zhu H, et al. (2014) Analysis of key genes and modules during the courses of traumatic brain injury with microarray technology. Genet Mol Res 13: 9220-9228. [Crossref]
- Kobori N, Clifton GL, Dash P (2002) Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res Mol Brain Res 104: 148–158. [Crossref]
- Tweedie D, Rachmany L, Kim DS, Rubovitch V, Lehrmann E, et al. (2016) Mild traumatic brain injury-induced hippocampal gene expressions: The identification of target cellular processes for drug development. J Neurosci Methods. [Crossref]
- Darkazalli A, Vied C, Badger CD, Levenson CW (2016) Human Mesenchymal Stem Cell Treatment Normalizes Cortical Gene Expression after Traumatic Brain Injury. J Neurotrauma. [Crossref]
- Kim CC, Nakamura MC, Hsieh CL, et al. (2016) Brain trauma elicits non-canonical macrophage activation states. J Neuroinflammation 13: 117. [Crossref]
- Merchant-Borna K, Lee H, Wang D, Bogner V, van Griensven M5, et al. (2016) Genome-Wide Changes in Peripheral Gene Expression following Sports-Related Concussion. J Neurotrauma 33: 1576-1585. [Crossref]
- Lončarević-Vasiljković N, Milanović D, Pešić V, Tešić V, Brkić M (2016) Dietary restriction suppresses apoptotic cell death, promotes Bcl-2 and Bcl-xl mRNA expression and increases the Bcl-2/Bax protein ratio in the rat cortex after cortical injury. Neurochem Int 96: 69-76. [Crossref]
- Dash PK, Hergenroeder GW, Jeter CB, Choi HA, Kobori N, et al., (2016) Traumatic Brain Injury Alters Methionine Metabolism: Implications for Pathophysiology. Front Syst Neurosci 10: 36. [Crossref]
- Cho YE, Latour LL, Kim H, Turtzo LC, Olivera A, et al. (2016) Older Age Results in Differential Gene Expression after Mild Traumatic Brain Injury and Is Linked to Imaging Differences at Acute Follow-up. Front Aging Neurosci 8: 168. [Crossref]
- Heinzelmann M, Reddy SY, French LM, Wang D, Lee H, et al. (2014) Military personnel with chronic symptoms following blast traumatic brain injury have differential expression of neuronal recovery and epidermal growth factor receptor genes. Front Neurol 5: 98. [Crossref]
- Stemper BD, Shah AS, Pintar FA, McCrea M, Kurpad SN, et al. (2015) Head rotational acceleration characteristics influence behavioral and diffusion tensor imaging outcomes following concussion. Ann Biomed Eng 43: 1071-1088. [Crossref]
- Jorge RE (2015) Mood disorders. Handb Clin Neurol 128: 613-631. [Crossref]
- Zgaljardic DJ, Seale GS, Schaefer LA, Temple RO, Foreman J, et al. (2015) Psychiatric Disease and Post-Acute Traumatic Brain Injury. J Neurotrauma 32: 1911-1925 [Crossref]
- Marsden WN1 (2013) Synaptic plasticity in depression: molecular, cellular and functional correlates. Prog Neuropsychopharmacol Biol Psychiatry 43: 168-184. [Crossref]
- Martins-de-Souza D, Guest PC, Harris LW, Vanattou-Saifoudine N, Webster MJ, et al. (2012) Identification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients. Transl Psychiatry 2: e87. [Crossref]
- Morries LD, Cassano P, Henderson TA (2015) Treatments for traumatic brain injury with emphasis on transcranial near-infrared laser phototherapy. Neuropsychiatr Dis Treat 11: 2159–2175. [Crossref]
- Xie LX, Calafat J, Janssen H, de la Iglesia-Vicente J, Mollinedo F (2010) Intracellular location of syntaxin 7 in human neutrophils. Immunol Lett 129: 72-77. [Crossref]
- Pan Y, Xia L (2015) Emerging roles of podoplanin in vascular development and homeostasis. Front Med 9: 421-430. [Crossref]
- Kokovay E, Wang Y, Kusek G, Wurster R, Lederman P, et al. (2012) VCAM1 is essential to maintain the structure of the SVZ niche and acts as an environmental sensor to regulate SVZ lineage progression. Cell Stem Cell 11: 220-230. [Crossref]
- Bramlett HM, Dietrich WD (2004) Pathophysiology of cerebral ischemia and brain trauma: similarities and differences. J Cereb Blood Flow Metab 24: 133-150. [Crossref]
- Günther E, Walter L (2001) The major histocompatibility complex of the rat (Rattus norvegicus). Immunogenetics 53: 520–542. [Crossref]
- Ethell IM, Ethell DW (2007) Matrix metalloproteinases in brain development and remodeling: synaptic functions and targets. J Neurosci Res 85: 2813-2823. [Crossref]
- Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG (2009) Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol 187: 733-747. [Crossref]



