Abstract
Ankylosing spondylitis (AS) is a common inflammatory rheumatic disease that affects the axial skeleton, causing characteristic inflammatory back pain, which can lead to structural and functional impairments and a decrease in quality of life. The cause of ankylosing spondylitis is unclear. There is increasing evidence that long (>200nt) non-coding RNAs (ncRNAs) have important regulatory functions in epigenetic regulation and involve in various kinds of disease. This study was to analysis the relationship between long ncRNAs and AS, further understand the basic biology of AS and identify new targets for therapeutics. We used microarray and real-time PCR to analysis the long ncRNAS in AS. In this study, we found differential expression hundreds of long ncRNAs in AS between patients and the controls through microarray analysis and quantitative real-time RT PCR confirmation, many ncRNAs surround or overlap protein-coding genes and can be predicted to function via a range of regulatory mechanisms. These ncRNAs may be new biomarkers, at the same time, let us understand the basic biology of AS and identify new targets for therapeutics.
Key words
Long non-coding RNA, microarray, Ankylosing spondylitis, epigenetic regulation, biomarkers
Introduction
Ankylosing spondylitis (AS) is a form of seronegative spondyloarthritis, chronic inflammatory and autoimmune disease. The main clinical features of AS is inflammatory back pain by affected the sacroiliac joints and spine [1]. Approximately 90% of AS patients are positive for HLA B27 and AS mainlly affects young people [2]. Many cytokines be involved in ankylosing spondylitis, such as Tumor necrosis factor-alpha (TNF α), IL-6 and so on [3]. Some studies displayed that AS are associated with inflammatory bowel disease and suggested the bacteria have central role in the pathogenesis of AS [4]. Despite intensive research, the cause of ankylosing spondylitis is unclear.
Long non-coding RNAs (lncRNAs) are a form of noncoding RNA and stranscripts longer than 200 nucleotides. LncRNAs are widely expressed in the mammalian and more and more increasing evidences that lncRNAs have important regulatory functions in gene transcription, post-transcriptional mRNA controls and epigenetic modifications [5-7]. People have focused on lncRNAs potential role in various human diseases, based on the steady accumulation of recognition that lncRNAs function. Some studies have revealed lncRNAs involved in human diseases, such as lncRNA TUG1 (necessary for retinal development) are upregulated in Huntington's disease (HD), while the brain-specific tumour-suppressor lncRNA MEG3 is downregulated [8]; lncRNA UCA1 regulated cell cycle through CREB via PI3K-AKT dependent pathway in bladder cancer[9], and so on.
Multiple lines of evidence increasingly link mutations and dysregulations of lncRNAs to a diverse number of human diseases. But until now, the possible correlation between changes in expression of lncRNAs and AS has not been studied. Through microarray analysis and quantitative real-time PCR confirmation, we find some lncRNAs associated with AS in this study. These lncRNAs deregulation may regulate neighboring protein-coding genes that are closely related to AS. This research provides an important foundation for future research on AS.
Materials and methods
Patients and controls
We studied blood of eleven patients with AS and eleven healthy volunteers. All patients consistent with the modified New York criteria 1984 for ankylosing spondylitis, they radiographic sacroiliitis grade ≥2 bilaterally, and present clearly one out of three Clinical criteria. All AS patients were recruited from the inpatient in the department of rheumatism in the 181th hospital in Guilin and were free of other autoimmune diseases. Age-, race-, and sex-matched healthy controls were recruited by advertising. None of the patients received immunosuppressive treatment and non-steroidal anti-inflammatory drugs at the time of the study. The control group got from health adult, all of health adult is voluntary, they were diagnosed with completely healthy and without any inflammatory. Patients and control samples were obtained with informed consent and approved by the Regional Ethics Committee of 181th Hospital of Guangzhou military area of PLA, Guilin.
Isolation of peripheral blood mononuclear cells
Blood samples were obtained from AS patients (n = 11) and from normal healthy donors (n = 11). The blood with Heparin tube (5 ml per subject) was diluted with equal volumes of phosphate-buffered saline. An equal volume of diluted blood was overlaid on Ficoll-Paque Plus in a 1:1 ratio and centrifuged at 800 g for 25 min at 22°C. Peripheral blood mononuclear cells (PBMCs) layer was harvested and washed with phosphate-buffered saline two times to remove plasma and Ficoll (Axis-shield, Norway). Then, these samples were stored at -80° C until be assayed.
RNA isolation and target labeling
Total RNAs was harvested using TRIzol (Invitrogen, USA) and the RNeasy kit (Qiagen, Germany) according to the manufacturer’s instructions. This extraction included a DNase digestion step. After passing RNA measurement quality control on the Nanodrop ND-1000 and denaturing gel electrophoresis, the RNA was used to synthesize double-stranded cDNA using the Superscript Double-Stranded cDNA Synthesis Kit (Invitrogen, USA). In the AS group, 11 samples of cDNA were labeled with Cy3-Random Nonamers; In the normal control group, 11 samples of cDNA were labeled with Cy3-Random Nonamers too. Then, the mixed double-stranded cDNAs of AS and normal control group were compared by individual hybridizing to a 12x135K LncRNA Expression microarray using the NimbleGen Hybridization System.
Microarray expression analysis
The microarray used is designed for the global profiling of long transcripts, including lncRNAs (long non-coding RNAs) and protein coding mRNAs. Each transcript is represented by 1-5 unique probes used to improve statistical confidence in the result. Probes for reference genes and negative probes are printed multiple times to ensure hybridization quality. 18,534 human lncRNAs were collected from the authoritative data sources including NCBI RefSeq, UCSC, RNAdb, lncRNAs from literatures, and UCRs. Sequences from these data sources are selected carefully using special strategies. Highly similar sequences and ncRNAs shorter than 200 bp are excluded. 18.847 protein coding genes from NCBI RefSeq are also present on this array. This enables the detection of mRNAs and lncRNAs in a single experiment.
Raw data were extracted as pair files by NimbleScan software (version 2.5). NimbleScan software’s implementation of RMA offers quantile normalization and background correction of data. Probe level normalization (*_norm_RMA.pair) files and Gene summary (*_RMA.calls) files were produced. The gene summary files were imported into Agilent GeneSpring Software (version11.0) for further analysis. Differentially expressed genes were identified through Fold-change screening.
Statistical analysis
Signal intensities for each spot were analyzed and calculated by Axon GenePix 4000B microarray scanner (Axon, USA), NimbleScan software (version 2.5, Nimblegen, USA) and Agilent GeneSpring Software (version11.0, Agilent, USA). Signal intensities for each spot were scanned and calculated by subtracting local background (based on the median intensity of the area surrounding each spot) from total intensities. An average value of the 1-5 spot replicates of each lncRNA was generated after data transformation (to convert any negative value to 0.01). Normalization was performed using a per-chip 50th percentile method that normalizes each chip to its median. This normalization allows the user to compare across chips. To highlight lncRNAs that characterize each group, a per-gene on median normalization was performed. This normalizes the expression of every lncRNA by its median across samples.
Pathway analysis and Gene Ontology (GO) term analysis by differentially expressed mRNAs
Pathway analysis is based on the latest KEGG (Kyoto Encyclopedia of Genes and Genomes) database. This analysis allows users to determine the biological pathways involved with their differentially expressed mRNAs.
GO analysis is a functional analysis that associates differentially expressed mRNAs with GO categories. The GO categories are derived from Gene Ontology (www.geneontology.org), which comprises three structured networks of defined terms used to describe gene product attributes.
The P-value denotes the significance of the pathway and the GO term enrichment in the differentially expressed mRNA list. The lower the P-value, the more significant the pathway and the GO Term (P-value <= 0.05 is recommended).
Quantitative real time RT-PCR
Total RNA extracted from various samples was 2-fold serially diluted in nuclease-free water. The diluted total RNA was used as a template for real-time RT-PCR. Data were collected in duplicate for each sample. Master mix without total RNA was prepared for all reactions with 24 μl being aliquoted into each reaction tube. 1 μl of the diluted total RNA was added to each reaction. Reactions were conducted using the Rotor-Gene 3000 Real-time PCR system (Corbett Research) with the following reaction profile: cDNA synthesis for 60 min at 37°C; pre-denaturation for 5 min at 95°C; and PCR amplification for 40 cycles with 10 sec at 95°C, 15 sec at 58°C, and 20 sec at 72°C. The PCR was followed by a melt curve analysis to determine the reaction specificity. Agarose gel electrophoresis was performed to confirm the size of PCR product. The mean Ct value was determined after the reaction to test the linearity of the GAPDH expression level.
These following primers (Primer sequences are listed in Table 1) were used to quantify ST6GAL1, DB065433, LCMT1, uc002dnw, CD3D, AK093775, PHC1 and DB483692 levels. GAPDH RNA was quantified as a control to normalize differences in total RNA levels.
Table 1. Oligonucleotide primer for cDNA amplication
|
Oligonucleotide
|
Sequence
|
|
GAPDH forward primer
|
5’GGGAAACTGTGGCGTGAT3’
|
|
GAPDH reverse primer
|
5’GAGTGGGTGTCGCTGTTGA3’
|
|
ST6GAL1 forward primer
|
5’GCATTCCGACAGCAGGACATA3’
|
|
ST6GAL1 reverse primer
|
5’AGAAGACAGAACCCCATAGACACA3’
|
|
DB065433 forward primer
|
5’CGCCGATTTTATTTGGAGAG3’
|
|
DB065433 reverse primer
|
5’CTCATGCCACATTGGGTTTT3’
|
|
LCMT1 forward primer
|
5’GCAACCAAAGGAGGAAATGAG3’
|
|
LCMT1 reverse primer
|
5’CAACAGGAACAGGAAGAGGAAC3’
|
|
uc002dnw forward primer
|
5’CGACAGCACTCTCCTCCTCA3’
|
|
uc002dnw reverse primer
|
5’TCCTCCCACTCCGCAAAC3’
|
|
CD3D forward primer
|
5’CTGGCTACCCTTCTCTCGCA3’
|
|
CD3D reverse primer
|
5’TGTTCCCACCGTTCCCTCT3’
|
|
AK093775 forward primer
|
5’CCATCAACGGCAGAAAGCA3’
|
|
AK093775 reverse primer
|
5’GCCAGGTCACCGAACTATCAG3’
|
|
PHC1 forward primer
|
5’GTCCCACTTCTGCTTCCGTTC3’
|
|
PHC1 reverse primer
|
5’GCTTTTCTTACTCCTGGTTCCCTA3’
|
|
DB483692 forward primer
|
5’CGCAGCAGAATGAATAGCCAG3’
|
|
DB483692 reverse primer
|
5’ACCGAGTAGAAGCAAAAGAGCAC3’
|
LncRNA relative to adjacent protein-coding genes analysis
LncRNA originated from complex transcriptional loci. We analyzed lncRNA relative to adjacent protein-coding genes and found that there were four groups. (1) cis-antisense lncRNAs were from the same genomic locus as their target transcript but from the opposite DNA strand. Cis-antisense lncRNAs form perfect or piece pairs with their target. (2) intronic lncRNAs were mapped within the intron of a protein-coding gene. (3) promoter-associated lncRNAs were defined when the lncRNA transcript across the protein-coding gene′s start site. (4) bi-directional lncRNAs and adjacent protein-coding gene whose transcriptions start sites directed away and are separated by less than 1000 base pairs.
Results
Differential expression of thousands of lncRNAs in AS
In this study, we performed fold-change filtering between the patients and the control samples from the experiment to identify differentially expressed lncRNAs in AS. The threshold we used to screen Up or Down regulated genes is fold-change >= 2.0 for mRNA transcripts and fold-change >= 2.0 for lncRNAs. After normalization and fold-change filtering, we found that 12.52% (2360 out of 18,847) of protein-coding transcripts in AS samples were significantly differentially expressed (fold-change >= 2.0) and 11.70% (2169 out of 18,534) of lncRNA transcripts were significantly differentially expressed (fold-change >= 2.0). There are 1836 up-regulated and 974 down-regulated in differentially expressed protein-coding transcripts; There are 556 up-regulated and 1613 down-regulated in differentially expressed lncRNA transcripts.
LncRNA relative to adjacent protein-coding genes analysis
Thought microarray chips screen, we found some lncRNAs and their adjacent protein-coding genes have closely relation in AS. They are ATP1A1 and uc001egg, ST6GAL1 and DB065433, LRP1 and uc009zpi, LCMT1 and uc002dnw, CD3D and AK093775, PHC1 and DB483692.
To confirm the microarray data, real-time PCR analyses were performed on the expression levels of lncRNAs and genes that mentioned above. Table 2, which summarizes and compares the real-time PCR and microarray results, confirms that the expression of lncRNAs and adjacent protein-coding genes that mentioned above and they have closely relation in AS. We analyzed lncRNA relative to adjacent protein-coding genes and found that there were four groups.
Table 2. Microarray and real-time PCR analyses of lncRNAs and adjacent protein-coding genes in blood of patients with AS.
|
LncRNAs or Genes
|
Microarray, fold changes
|
Real-time PCR, fold changes
|
|
ST6GAL1
|
7.04
|
5.13
|
|
DB065433
|
-17.69
|
0.19
|
|
LCMT1
|
4.15
|
2.81
|
|
uc002dnw
|
-7.14
|
0.31
|
|
CD3D
|
7.08
|
5.82
|
|
AK093775
|
-9.50
|
0.34
|
|
PHC1
|
12.74
|
24.68
|
|
DB483692
|
-5.86
|
0.13
|
Cis-antisense lncRNAs
The cis-regulatory impact on cis-antisense genes is of functional importance [10]. Current evidence has shown a variety of regulatory roles for cis-antisense lncRNAs like reprogramming of chromatin, RNA interference (RNAi), alternative splicing, genomic imprinting, and X-chromosome inactivation [11].
We found an lncRNA (uc001egg, ATP1A1-ncRNA) that was identified as being antisense to the ATPase, Na+/K+ transporting, alpha 1 polypeptide (ATP1A1) (Figure 1A). Na/K-ATPase acts as a signal transducer that regulates the function of protein kinases and activation of several other cascades, including p42/44 and p38 MAPKs, et al. [12]. ATP1A1 encodes a protein belongs to the family of P-type cation transport ATPases, and to the subfamily of Na+/K+ -ATPases. Some evidence suggests that during the lymphocyte transition from resting stage to proliferation, long-term activation of Na,K-ATPase pump is due to the enhanced expression of Na,K-ATPase protein and mRNA, the level of Na,K-ATPase alpha1-subunit mRNAs significant increase [13]. The antisense ncRNA, which we named ATP1A1-ncRNA, overlaps with the ATP1A1 in AS (Figure 1A). We found increased expression of ATP1A1 was in AS patient samples, and the decreased expression of ATP1A1-ncRNA. We inferred that uc001egg may negatively regulate the ATP1A1 gene.

Figure 1. LncRNA lies opposite to associated protein-coding gene. A. ATP1A1 and ATP1A1-ncRNA (uc001egg) genomic locus. B. ST6GAL1 and ST6GAL1-ncRNA (DB065433) genomic
2021 Copyright OAT. All rights reserv
locus.
We also identified another antisense ncRNA (DB065433, ST6GAL1-ncRNA) that lies opposite of ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (ST6GAL1) gene in a highly complex locus (Fig.1B). The encoded protein is a type II membrane protein that catalyzes the transfer of sialic acid from CMP-sialic acid to galactose-containing substrates. Galectins are β-galactoside-binding lectins that regulate diverse cell behaviors, including adhesion, migration, proliferation, and apoptosis, some evidence suggesting that ST6Gal-I activity serves as an "off switch" for galectin function [14]. Macrophages play a central role in innate immunity, other evidence suggesting ST6Gal-I can regulate macrophage apoptosis [15]. In this study, we found that ST6GAL1 gene expression was highly up-regulated, but the expression of its antisense ncRNA, which we named ST6GAL1-ncRNA, was drastically down-regulated. Quantitative Real time RT-PCR experimental results validate these observations. DB065433 may negatively regulate the ST6GAL1 gene.
Intronic lncRNAs
At complex loci, we observed a large number of lncRNAs within the introns of protein-coding genes. LncRNAs from introns have diverse regulatory functions, including being the precursors of shorter RNAs, protein-coding RNA stabilization, the control of gene expression, and the regulation of alternative splicing in protein-coding RNA [16,17]. For example, we identified an ncRNA (uc009zpi, LRP1-ncRNA) located within the introns of low density lipoprotein receptor-related protein 1 (LRP1) (Figure 2A). The protein encoded by this gene is an endocytic receptor involved in several cellular processes, including intracellular signaling, lipid homeostasis, and clearance of apoptotic cells. LRP1 plays an important role in regulating immune responses, inflammation and phagocytosis [18]. In our study, we observed up-regulation in LRP1 gene expression. Expression of its antisense ncRNA, which we named LRP1-ncRNA, was up-regulated. This observation suggests that LRP1 and LRP1-ncRNA are both associated with this disease, but might be independently regulated.
We also identified another intronic ncRNA (uc002dnw, LCMT1-ncRNA) that was located within the introns of leucine carboxyl methyltransferase 1 (LCMT1) (Figure 2B). LCMT1 catalyzes the methylation of the carboxyl group of the C-terminal leucine residue (leu309) of the catalytic subunit of protein phosphatase-2A. These are some evidences suggests that LCMT-1 is important for normal progression through mitosis and cell survival [19]. In this study, we found that LCMT-1 gene expression was up-regulated, but the expression of its intronic ncRNA, which we named LCMT-1-ncRNA, was down-regulated. Quantitative Real time RT-PCR experimental results validate these observations. From these data we inferred that uc002dnw may negatively regulate the LCMT-1 gene.
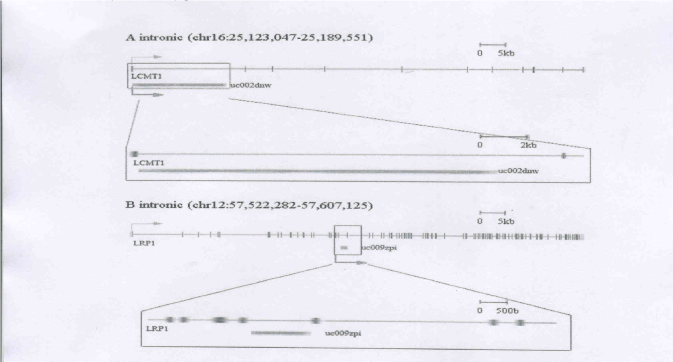
Figure 2. LncRNA within the introns of associated protein-coding gene. A. LCMT1 and LCMT1-ncRNA (uc002dnw) genomic locus. B. LRP1 and LRP1-ncRNA (uc009zpi)
genomic locus.
Promoter-associated lncRNAs
Transcription of lncRNAs interact with downstream promoter region of protein-coding genes, these lncRNAs help regulate mRNA expression [20]. For example, a lncRNA (AK093775) across the transcription start site of CD3D molecule, delta (CD3D), which we named CD3D-ncRNA (Figure 3A). Gammadelta T cells is a link between innate and adaptive immune responses [21]. The CD3D gene encoded protein is part of the T-cell receptor/CD3 complex (TCR/CD3 complex), and while CD3D mutation would affect all T cells, thus causing severe combined immunodeficiency disease (SCID) [22]. Has research found that the CD3D gene is up-regulated in primary Sjögren's syndrome patients (Sjögren's syndrome is a systemic autoimmune disease). Interestingly, we observed that CD3D gene expression was up-regulated, but the expression of CD3D-ncRNA, was down-regulated. Quantitative Real time RT-PCR experimental results validate these observations. From these data we inferred that AK093775 may negatively regulate the CD3D gene.
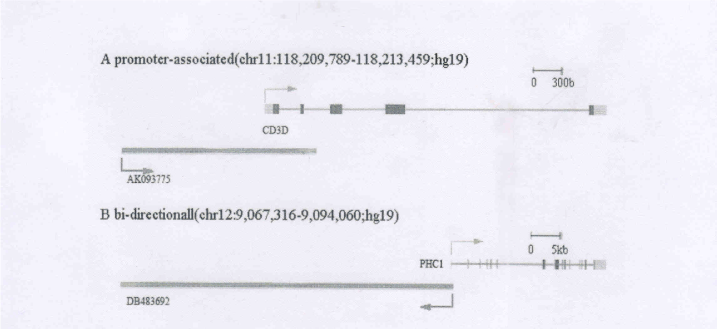
Figure 3. A. LncRNAs cross the promoter of associated protein-coding genes, CD3D and CD3D-ncRNA (AK093775) genomic locus. B. Bi-directional lncRNAs and adjacent proteincoding
gene, PHC1 and PHC1-ncRNA (DB483692).
Bi-directional lncRNAs
Bi-directional lncRNAs and adjacent protein-coding gene whose transcriptions start sites directed away and are separated by less than 1000 base pairs. Promoter sequences between divergently transcribed lncRNA and gene pairs that initiate transcription in both directions. Bidirectional promoters have received considerable attention because of their ability to regulate two downstream genes [23]. Increasing evidence indicates that non-coding transcription at promoters influences the expression of protein-coding genes, revealing a new layer of transcriptional regulation [24]. For example, we identified an lncRNA (DB483692,PHC1-ncRNA) and polyhomeotic homolog 1(PHC1)gene whose transcription start sites are neighboring and directed away from each other. They are separated by less than 1000 base pairs (Figure 3B). PHC1 gene is a human homolog of the Drosophila polyhomeotic gene, which is a member of the Polycomb group of genes, and can mediate polycomb to suppress repression of Hox genes. In the immune system, some results suggest polycomb group proteins play an important regulatory role [25]. In this study, we found that PHC1 gene expression was up-regulated, but the expression of lncRNA, which we named PHC1-ncRNA, was down-regulated. Quantitative Real time RT-PCR experimental results validate these observations. From these data we inferred that DB483692 may negatively regulate the PHC1 gene.
Discussion
In this study, we focus on the genomic context of lncRNAs. We aim to analyze complex transcriptional loci that include lncRNAs and their associated protein-coding genes. Some evidence suggests that lncRNAs can regulate gene expression through epigenetic regulation of chromatin modification, transcription and post-transcriptional processing [7].
LncRNAs are transcribed in complex intergenic, overlapping and antisense patterns relative to adjacent protein-coding genes, suggesting that many lncRNAs regulate the expression of these genes. In many cases, the long noncoding RNAs themselves serve key regulatory roles that were assumed previously to be reserved for proteins, such as transcriptional regulation, genomic imprinting, and protein transport [7-9]. In addition, many long noncoding RNAs are processed to yield small RNAs or, conversely, modulate how other RNAs are processed [17]. The link between lncRNAs and some human diseases has been proven [8,9]. However, at the present time the function of only a minority of long ncRNAs is documented. So in this study, we were therefore interested in studying the relationship between AS and long ncRNAs in the hope of gaining new insights into the pathophysiological mechanism of AS, which has not been studied before.
In summary, we presented here an analysis of lncRNAs expression in control and AS PBMC samples using microarray. As analyzed above, these lncRNAs affect the expression of their associated protein-coding genes. The malfunction of regulation in the network of lncRNAs may be a possible mechanism for the provocation of AS. Our works indicates that lncRNAs are potential pathophysiological mechanism and probable factors involved in the pathogenesis of AS. Nevertheless, it is important to note that the findings described in this article are merely a starting point for the study of lncRNAs in AS, and much exciting work lies ahead to functionally characterize the identified lncRNAs. This novel and powerful technology should provide additional opportunities to advance our understanding of the pathophysiological mechanism of AS.
Conflicts of interest
The authors have no financial conflict of interest. This study was supported by the Guangxi of China Natural Science Fundation (2011GXNSFB018105)
Acknowledgment
We would like to thank the patients and healthy volunteers who participated in this study.
References
- Tam LS, Gu J, Yu D (2010) Pathogenesis of ankylosing spondylitis. Nat Rev Rheumatol 6: 399-405. [Crossref]
- Feldtkeller E, Khan MA, van der Heijde D, van der Linden S, Braun J (2003) Age at disease onset and diagnosis delay in HLA-B27 negative vs. positive patients with ankylosing spondylitis. Rheumatol Int 23: 61-66. [Crossref]
- Bal A, Unlu E, Bahar G, Aydog E, Eksioglu E, et al. (2007) Comparison of serum IL-1 beta, sIL-2R, IL-6, and TNF-alpha levels with disease activity parameters in ankylosing spondylitis. Clin Rheumatol 26: 211-215. [Crossref]
- Mundwiler ML, Mei L, Landers CJ, Reveille JD, Targan S, et al. (2009) Inflammatory bowel disease serologies in ankylosing spondylitis patients: a pilot study. Arthritis Res Ther 11: R177. [Crossref]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155-159. [Crossref]
- Morris KV, Vogt PK (2010) Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle 9: 2544-2547. [Crossref]
- Morris KV (2009) Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 4: 296-301. [Crossref]
- Johnson R (2012) Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis 46: 245-254. [Crossref]
- Yang C, Li X, Wang Y, Zhao L, Chen W (2012) Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene 496: 8-16. [Crossref]
- Engström PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, et al. (2006) Complex Loci in human and mouse genomes. PLoS Genet 2: e47. [Crossref]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071-1076. [Crossref]
- Sheik Mohamed J Gaughwin PM, Lim B, Robson P, Lipovich L (2010) Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324-337. [Crossref]
- Xie Z1 (2003) Molecular mechanisms of Na/K-ATPase-mediated signal transduction. Ann N Y Acad Sci 986: 497-503. [Crossref]
- Karitskaya I, Aksenov N, Vassilieva I, Zenin V, Marakhova I (2010) Long-term regulation of Na,K-ATPase pump during T-cell proliferation. Pflugers Arch 460: 777-789. [Crossref]
- Zhuo Y, Bellis SL (2011) Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem 286: 5935-5941. [Crossref]
- Liu Z, Swindall AF, Kesterson RA, Schoeb TR, Bullard DC, et al. (2011) ST6Gal-I regulates macrophage apoptosis via α2-6 sialylation of the TNFR1 death receptor. J Biol Chem 286: 39654-39662. [Crossref]
- Li SC, Tang P, Lin WC (2007) Intronic microRNA: discovery and biological implications. DNA Cell Biol 26: 195-207. [Crossref]
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, et al. (2007) Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 8: R43. [Crossref]
- Stanevich V, Jiang L, Satyshur KA, Li Y, Jeffrey PD, et al. (2011) The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol Cell 41: 331-342. [Crossref]
- Abarrategui I, Krangel MS (2007) Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. EMBO J 26: 4380-4390. [Crossref]
- Beetz S, Wesch D, Marischen L, Welte S, Oberg HH, et al. (2008) Innate immune functions of human gammadelta T cells. Immunobiology 213: 173-182. [Crossref]
- Gil J, Busto EM, Garcillán B, Chean C, García-Rodríguez MC, et al. (2011) A leaky mutation in CD3D differentially affects αβ and γδ T cells and leads to a Tαβ-Tγδ+B+NK+ human SCID. J Clin Invest 121: 3872-3876. [Crossref]
- Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, et al. (2007) Transcription factor binding and modified histones in human bidirectional promoters. Genome Res 17: 818-827. [Crossref]
- Wei W, Pelechano V, Järvelin AI, Steinmetz LM (2011) Functional consequences of bidirectional promoters. Trends Genet 27: 267-276. [Crossref]
- Suzuki A, Iwamura C, Shinoda K, Tumes DJ, Kimura MY, et al. (2010) Polycomb group gene product Ring1B regulates Th2-driven airway inflammation through the inhibition of Bim-mediated apoptosis of effector Th2 cells in the lung. J Immunol 184: 4510-4520. [Crossref]



