The purpose of this study was to assess and explain the relationship between molecular changes occurring in squamous cell laryngeal carcinoma. Furthermore, we were revealed the role of FHIT gene response in the development and progression of squamous cell laryngeal carcinoma, and genes based exons 7,8 and 9 in tumour stage and histopathologic grade. In this study was used 50 blood samples for control and 26 patients with laryngeal squamous cell carcinoma. Briefly, for this purpose, we primarily aimed researching or evaluating exon 7 (C/T), exon 8 (T/C) and exon 9 (A/G) homozygous deletions of FHIT gene exons. Mutations were examined with polymerase chain reaction - single-strand conformation polymorphism (PCR-SSCP) and RT-PCR based HRM technigues. At the end of the HRM analysis, was observed a significant mutation in the groups of patients. L4 and L19 in the number and before the results of SSCP individuals identified as benign according to the results of HRM. In this study, exon 7 and 9 healthy individuals and squamous cell laryngeal carcinoma were observed any difference between individuals in terms of mutation. The pathology of the case was low differentiation. But in addition to this findings, the present findings highlight that the FHIT gene partially related to target for analysis in case with laryngeal squamous cell carcinoma. According to results of analyses, we idendified a link in between the exon 8 and exon 9 of FHIT gene.
FHIT gene, Squamous cell carcinoma, PCR-SSCP, HRM
SCC: squamous cell carcinoma; PCR-SSCP: polymerase chain reaction - single-strand conformation polymorphism; HRM: high-resolution melting.
Squamous Cell Carcinoma (SCC) represents more than 60 percent of all head and neck cancers. In Turkey, SCC of the head and neck comprises about 4 percent of all malignancies [1]. Since multiple specific genetic targets and their cancer-related alterations have been identified, it may be possible to use sophisticated and sensitive molecular assays to detect these abnormalities early in tumor progression. By detecting alterations early in the progression cycle, clinicians may be able to treat patients at a time when cure may be more attainable with current treatment protocols. The vast majority of tumors are squamous cell carcinomas [2]. Due to the important physiologic functions of the larynx, advanced laryngeal lesions are associated with significant morbidity and mortality for the patient and increased financial costs for society [3]. In addition, the disease can be identified early treatment of existing facilities can be provided with opportunities for long life. Late in the disease the patient's chance of survival decreases known and more radical methods of treatment are required [4-6]. Epidemiological studies conducted in our country were found between mean age 55-59 [7]. Molecular basis of larynx cancer has recently been investigated. Larynx cancers transformation of normal cells into cancer cells requires a multistage process. Tumor suppressor genes are normal genes that slow down cell division, repair DNA mistakes, or tell cells when to die (a process known as apoptosis or programmed cell death). When tumor suppressor genes don't work properly, cells can grow out of control, which can lead to cancer. FHIT (Fragile Histidine Triad Gene) was recently attracted attention in the genetic changes. DNA replication and cell cycle control play a role in protein-encoding gene is a tumor suppressor [7]. Through investigation of homozygous deletions in common human cancers, including lung cancer, The FHIT gene at the common fragile site at chromosome band 3p14.2 [8]. The results of the investigation of human tumors indicate that the FHIT gene is altered by mutations, predominantly deletions, in human cancer and that such alterations are common in epithelial cancers, particularly those resulting from exposure to environmental carcinogens, such as lung, larynx, head and neck, esophageal, stomach, pancreatic, and cervical cancer [9]. The FHIT, that is inactivated in epithelial tumors, particularly in tumors resulting from exposure to environmental carcinogens. In some tumors, particularly those associated with environmental carcinogens, alterations in the FHIT gene occur quite early in the development of cancer [10,11]. The purpose of this study, in the formation and progression of laryngeal squamous cell carcinoma reveal the role of FHIT gene, as well as the relationship between molecular changes occurring to explain. Because FHIT gene encodes carcinogen induced damage can lead to deletions.
This study includes 26 patients (3 females and 23 males) who applied to Cumhuriyet University of Ear Nose and Throat Department with diagnosis of larynx cancer without palpable neck nodes between 2009-2011 and healthy Turkish volunteers with their signed informed consents and patient's tumor tissue as controls any family history in the peripheral venous blood of 50 healthy individuals without disease were included.
Formalin-fixed and paraffin-embedded tissue was obtained from the archives of the Department of Pathology, University of Cumhuriyet, Sivas, Turkey. Representative stained sections of all patients were examined histopathologically by an experienced pathologist. All tumor samples selected for extraction contained mostly tumor cells. Genomic DNA from paired tumor and its corresponding normal tissue was extracted from serial unstained sections.
The DNA used for polymorphic analysis was isolated from the biopsy samples of patients with cancers by using DNA isolation kit purchased from nucleic acid isolation kit (Oiagen, Germany) the manufacturer’s instructions. Isolated DNA was stored at -20°C till use. The control group consisted of healthy unrelated volunteers without a medical history of cancer or other chronic diseases. All patients and controls were of Turkish population.
For Polymerase Chain Reaction (PCR) application (Biotechnology Research Laboratory, Turkey belonging Dr. H.C.V.), increasing concentrations of extracted DNA of each specimen was tested to find out the optimum dose that resulted in good amplicon product. Exon 7,8 and Exon 9 of the FHIT gene were amplified using primer designed according to Risinger et al. [12]. Each primer pairs of the selected exons was used for mutation detection of FHIT following the PCR for the single-strand conformation polymorphism (PCR-SSCP). Tm values or annealing Tm and primers (Table 1) used for each
FHIT exon were as follows: 4 µl larynx tissue genomic DNA was amplified in a mixture composed of 5 μl 10XPCR Taq buffer (pH 8.8), 2 mM MgCl2, and 10 mM dNTPs (dGTP, dATP, dTTP, dCTP) at each, 0.5 mM of each primer, and 0.3 U DreamTaq polymerase (Advanced Biotechnologies Ltd., Fermantase Life Science). Amplification was submitted to polymerase chain reaction for exon 7, 8, 9; Initial denaturation at 95°C 4 min, denaturation at 95°C 15 sec, annealing Tm at 57,5°C 30 sec, extension at 72°C 20 sec, total 35 cycles and final extension at 72°C 4 min. PCR was done on genomic DNA extracted from whole blood and parafine-embedded tissue samples using suitable primers to amplify these loci.
PCR primers:Exons 7,8 and 9 exons of the FHIT gene were used for mutation analysis. The oligonucleotide primers used for PCR were selected from ATQ–GenBank (Giagen Firm, GERMANY). The sequences of these exons and the annealing temperatures were listed in (Table 1).
Table 1. The sequences of these exons and the annealing temperatures
Locus |
Primer |
Tm |
PCR product |
FHIT-exon 7 |
Forward 5’-TGGTCCCCATGAGAATACTATAAATTAACA-3’
Reverse 3’-TTACGGCTCTAACACTGAGGGTCTCTCTGA-5’
|
60°C |
304 bp
|
FHIT-exon 8 |
Forward 5’-GAGTAATTGGGCTTCATGAGAGCATCACT-3’
Reverse 3’- AGGTTGATGTCATCCCACCGACAGT -5’
|
63°C |
224 bp |
FHIT-exon 9 |
Forward 5’-TTCTCCAAAGCTCCAGAAACATGACAAGGA-3’
Reverse 3’- GTCTTTACCTGTGTCACTGAAAGTAGACCC-5’
|
58.5°C |
119bp |
Eight percent neutral polyacrylamide gel electrophoresis was performed as previously described (Rohr et al.,2005; Eyzaguirre et al.,1999). In brief, 3 mL 40% acrylamide solution, 3 mL 5XTBE solution, 3 mL 50% glycerin, 6 mL ddH2O, 75 μL 10% ammonium presulfate, 7 μL TEMED, were blended adequately and poured into the gel, then concreted for 1 h at room temperature. Four μL PCR products and 6 μL formamide sample were mixed. The mixture was centrifuged for 15 s, denatured at 95 oC for 10 min, bathed in ice for 10 min, put on an 8% neutral polyacrylamide gel and electrophoresed with 1XTBE buffer for 8 h at 300 V. The fixation solution was infused into a flat utensil, into which gel was immerged, vibrated for 10 min, and washed 3 times (2 min each time) with ddH2O. The gel was immerged into a staining solution, vibrated for 10 min, washed 3 times (20 s each time) with ddH2O. The gel was then immerged into a display solution, vibrated until the sample signal became brown and the background became transparent yellow and rinsed with tap water to stop display. The staining results were observed and photographs were taken. According to the PCR-SSCP results of genome DNA, the difference in the single strand strip number and electrophoresis transference location, also known as the mobility shift, was considered PCR-SSCP positive.
In this case, we propose an extension of HRM analysis for identification of the similarities and dissimilarities in DNA using the sequence matching and genotype auto-call functions that uses the changes in the melting profiles of amplicon which can be in two forms: a shift in melting temperature and an obvious difference in the shape of the melt curve. HRM analysis allows for sequence matching and auto-calling of genotypes for unknown samples with appropriate references. Results can be viewed as a normalized melt plot, difference plots and auto-called genotypes with confidence values (%). Normalization provides the selection of suitable samples and analysis range with the basic representation of the different genotypes based on curve shifting (for homozygotes) and curve shape change (for heterozygotes). Difference plots are an aid to visual interpretation.
HRM analysis results, mutated patient groups subjected to DNA sequence analysis was performed mutation identification. Larynx cancer tissues obtained from patients genotype distribution of the statistical evaluation is made as to whether changes in this gene was determined. HRM experiments generate DNA melt curve profiles that are both specific and sensitive enough to distinguish nucleic acid species based on small sequence differences enabling, mutation. Both melting temperature shifts and curve shape can be used to identify sequence differences. Homozygous allelic/sequence variants are typically characterized by the temperature (x-axis) shift observed in an HRM melt curve, whereas heterozygotes are commonly characterized by a change in melt curve shape generated from base-pairing mismatches as a result of destabilized heteroduplex annealing between some of the wild-type and variant strands. HRM data are often plotted as difference curves to visually magnify differences between the melt profiles of different clusters within the same genotype. HRM analysis results, mutated patient groups subjected to DNA sequence analysis was performed mutation identification. Larynx cancer tissues obtained from patients genotype distribution of the statistical evaluation is made as to whether changes in this gene was determined. After each real-time PCR assay and HRM analysis were performed on the 76 samples. The results of the HRM analysis of the Sybr GreenER assay are shown in (Figures 1-8). The Sybr GreenER assay exhibits a melting pattern that begins its separation at approximately 77°C and ends at approximately 80.5°C. In this HRM study, 50 blood samples were used as controls. Larynx cancer, treated samples from 26 individuals were used. These examples has been studied in 7,8 and 9 exons of FHIT in the sense of mutation.
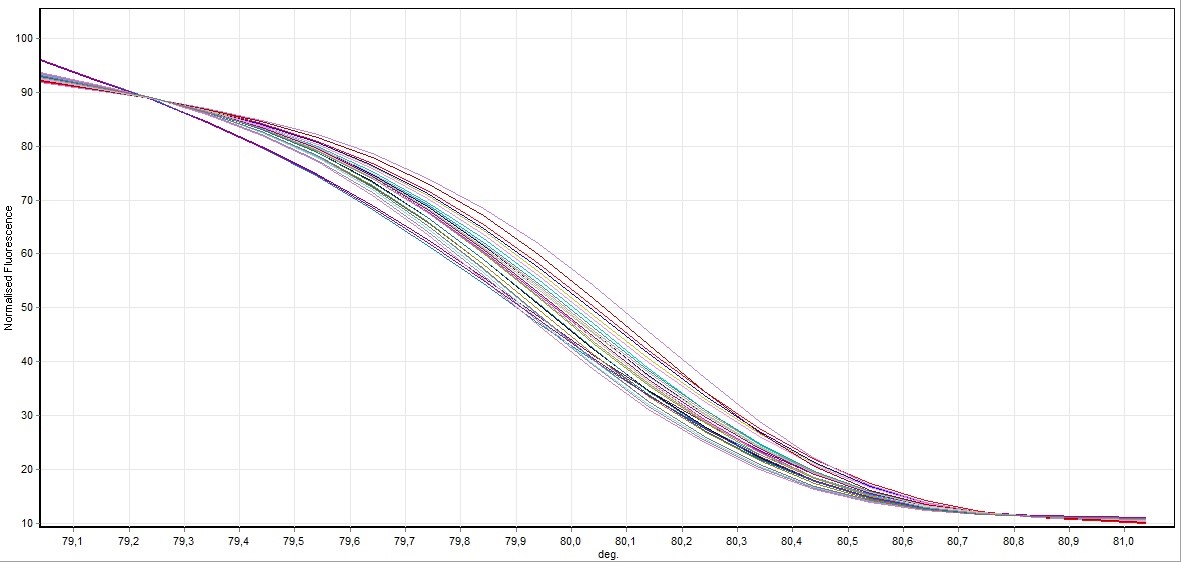
Figure 1. Direct visualization HRM analysis of FHIT gene exon 7 in all samples (control and cancer case)
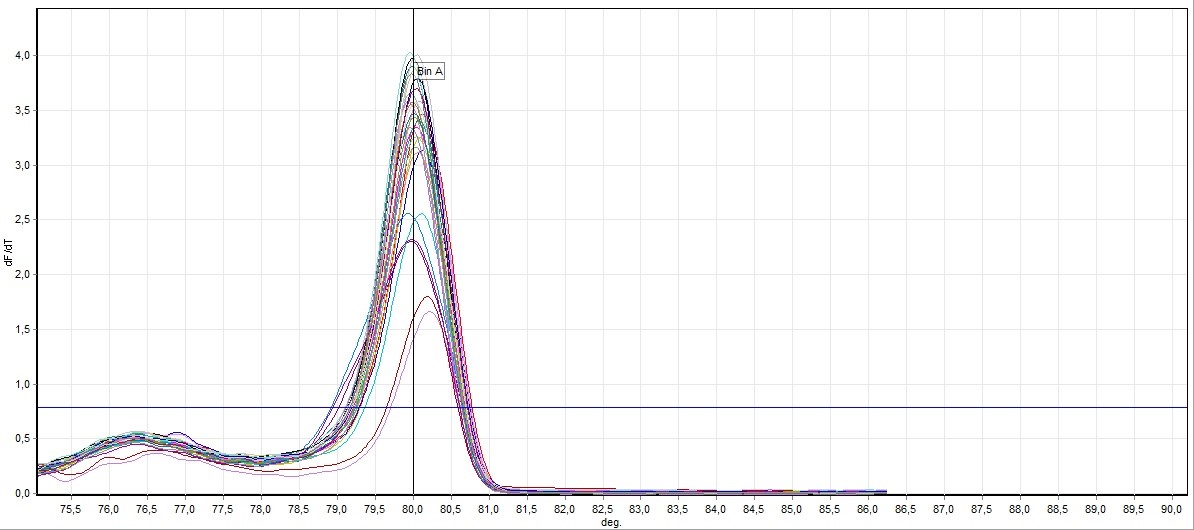
Figure 2. General screening of melting curve analysis of exon 7 in FHIT gene
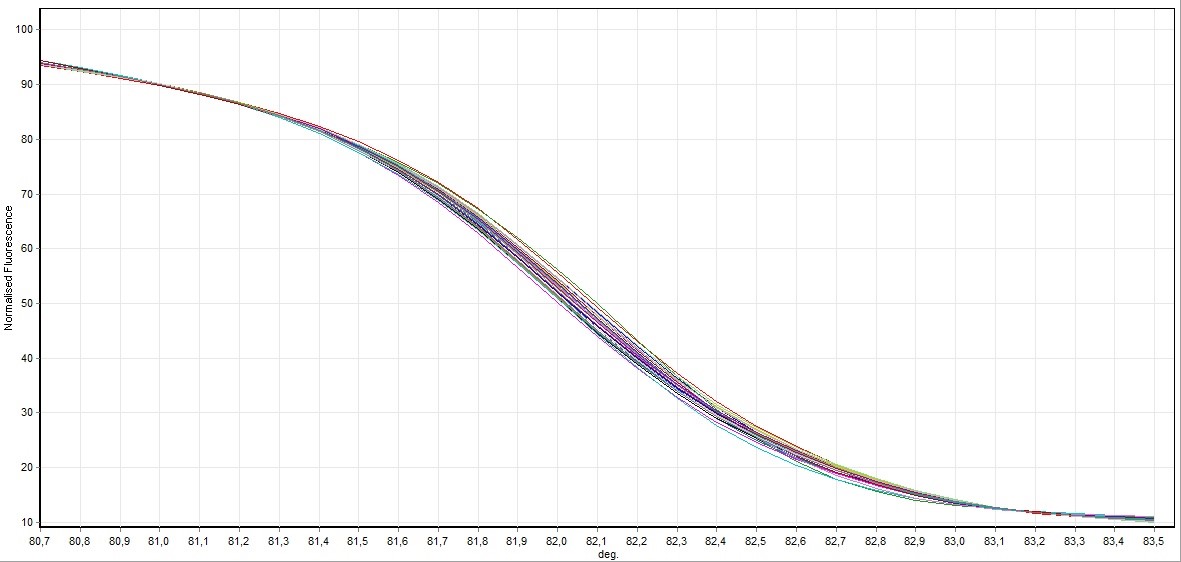
Figure 3. Direct visualization HRM analysis of FHIT gene exon 9 in all samples
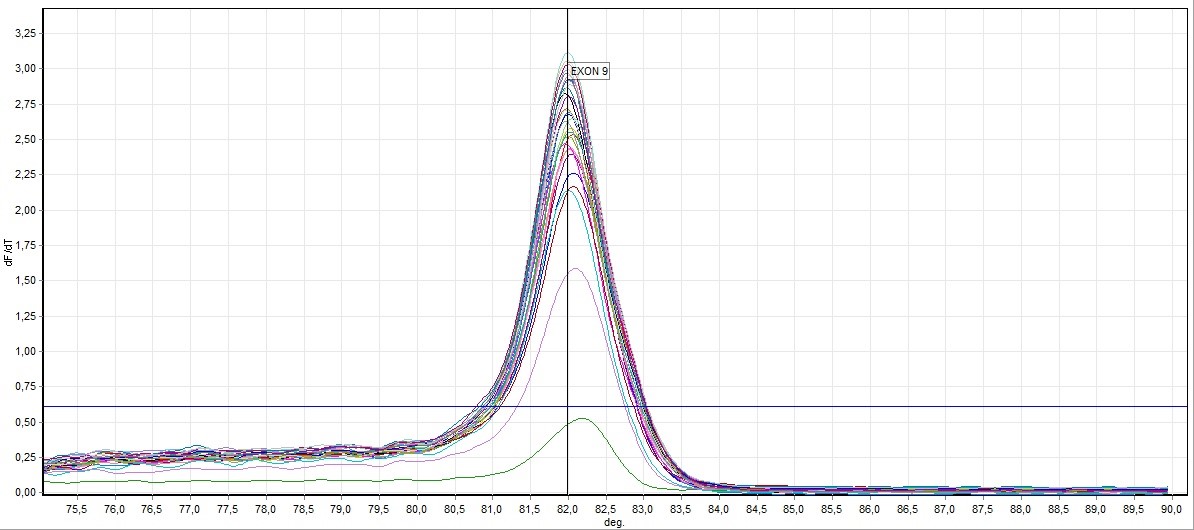
Figure 4. General screening of melting curve analysis of exon 9 in FHIT gene
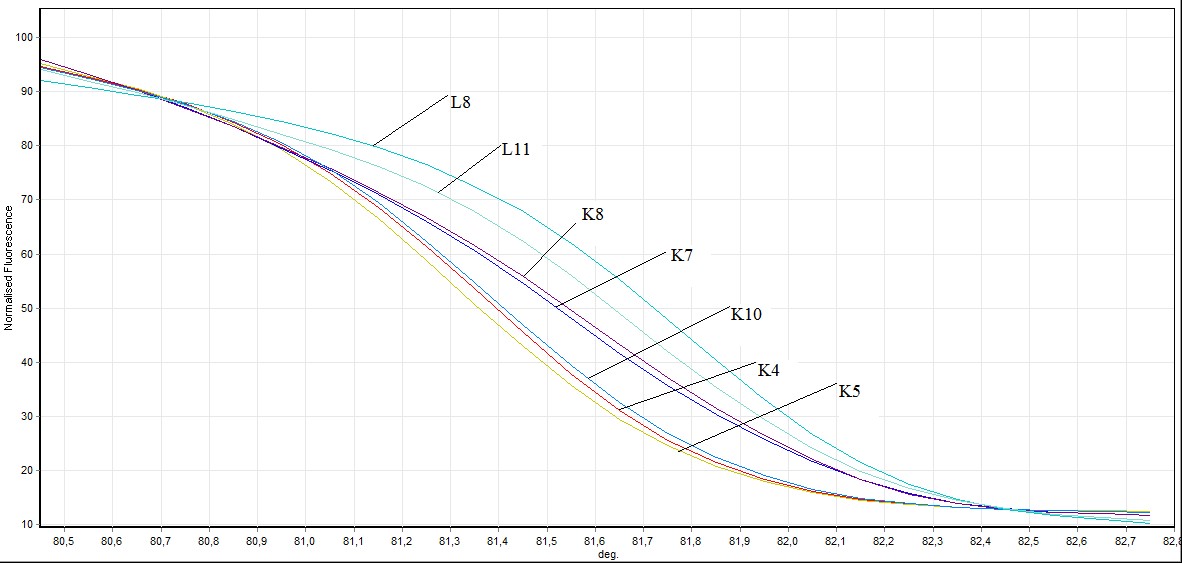
Figure 5. Direct visualization HRM analysis of FHIT gene exon 8 in mutated individuals and healthy individuals (L: mutated individuals K: healthy individuals)
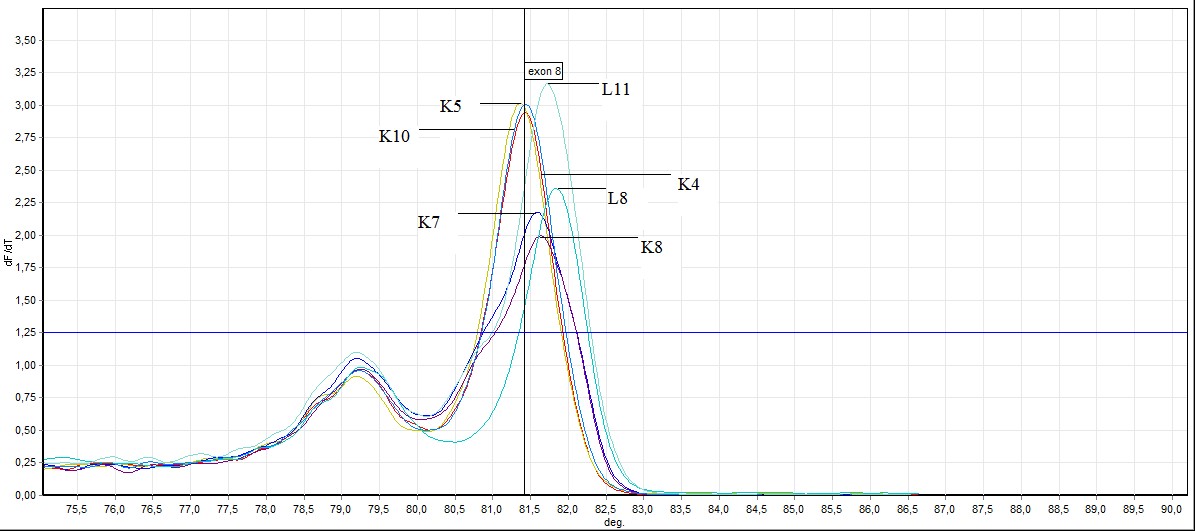
Figure 6. General screening of melting curve analysis of exon 8 in FHIT gene sequence (L: mutated individuals, K: healthy individuals)
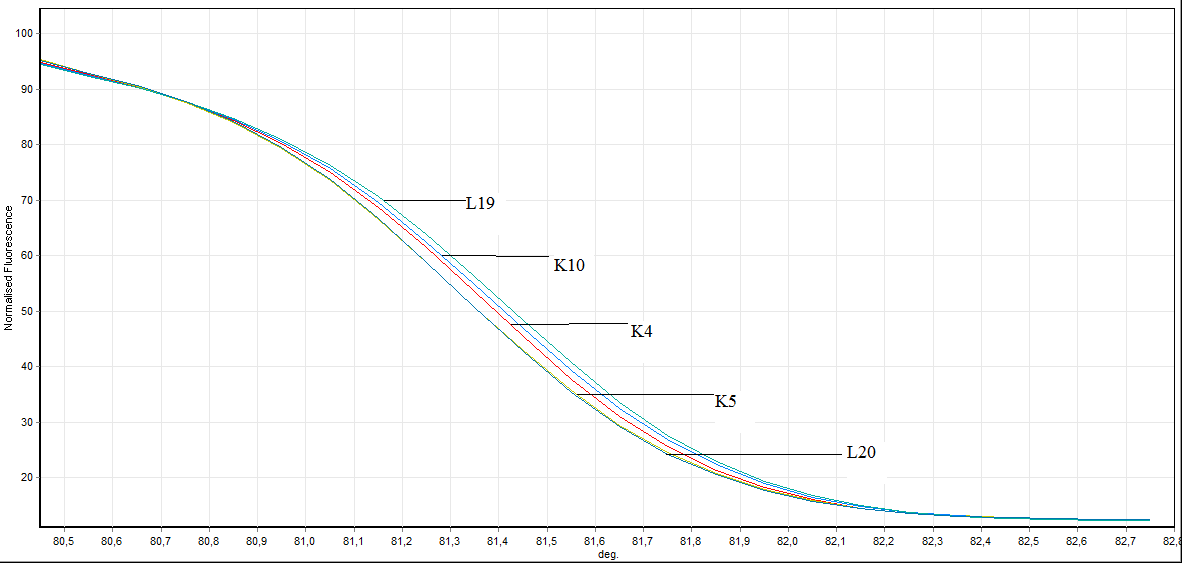
Figure 7. Direct visualization HRM analysis of FHIT gene exon 8 in bening individuals and healthy individuals (L: bening individuals K: healthy individuals)
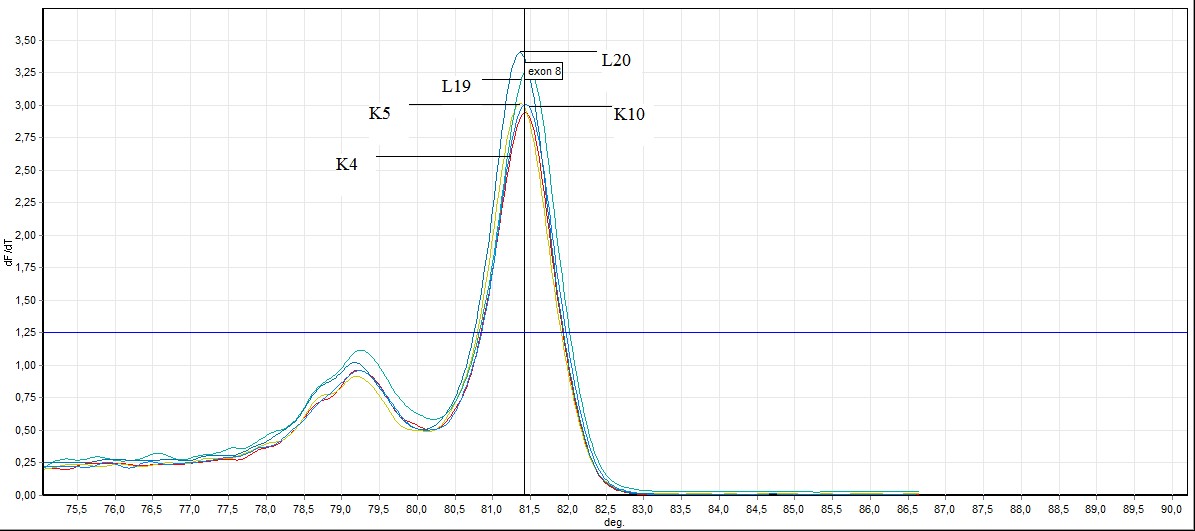
Figure 8. General screening of melting curve analysis of exon 8 in FHIT gene sequence in bening and healthy individuals (L: bening individuals, K: healthy individuals)
In the exon 8: in exon 8, 2 out of 26 samples taken from individuals (L8 ve L11) mutation, 3 individuals (L4, L19, L20 ) benign observed no difference was observed in other samples. Showing benign individuals: L19, L20 and L4
L4 ve L19 number of individuals, SSCP results and to the results of previously conducted by HRM has been identified as benign. In this study we HRM having this number of individuals has been identified as benign.
The molecular events which induce laryngeal tumorigenesis, especially laryngeal carcinogenesis, are not well characterized. Proto-oncogenes seem to be the target of the risk factors (cigarette smoking, alcohol abuse, ionizing radiation and human papillomavirus infection) that are commonly considered to be associated with laryngeal squamous cell carcinoma. Several tumor suppressor genes have been shown to play important roles in human tumors, including head and neck cancers. Head and neck cancers include malignancies arising from different anatomical sites within the upper aero-digestive tract. Head and neck cancers are characterized by heterogeneous histology. The majority carcinomas that arise from squamous cell epithelia are head and neck squamous cell carcinomas (HNSCC), while other cancer types that can occur in the head and neck include thyroid cancer, malignant salivary gland tumors, lymphomas and sarcomas. HNSCC include cancers of the oral cavity, larynx, pharynx (oropharynx, hypopharynx, nasopharynx), and esophagus [13-19]. Report on the role FHIT in the head and neck cancer are not uniform [8,20,21,22]. Loss of chromosome region 3p is a common early genetic event in HNSCC [23,24,25]. Genetic alterations at 3p that are common in HNSCC and OSCC contains tumor suppressor genes Fragile histidine triad (FHIT) gene mapped at 3p14, Ras association family (RASSF1A) gene, mapped at 3p21, and Retinoic acid receptor B2 gene (RARB2), mapped to chromosome region 3p24 [22,23,25-31] Amplification of 11q13 leads to overexpression of cyclin D1 [32], detected in 30–60% of HNSCC and 40% of cases of oral squamous dysplasia [33]. investigated the expression of FHIT in the squamous cell carcinomas of tongue by immunochemistry. They reported that loss of FHIT expression was seen in 68% of patients. They also emphasized the loss of FHIT as a negative prognostic indicator for clinical outcome [33-38 22]. The FHIT locus is involved in translocations and deletions in some fraction of many types of cancer, likely due to the recombinogenicity of the fragile region within FHIT and subsequent selective growth or survival advantage of cells with reduced FHIT protein expression. Deletions of the short arm of chromosome 3 are also observed in a variety of other common tumors, such as breast cancer, head and neck cancer, gastrointestinal cancer, esophageal cancer, and cervical cancer [11]. To identify the gene or genes on chromosome 3p that may be involved in the initial steps in the development of these tumors, it was first necessary to define the chromosomal location of these genes [10,39] The results of the investigation of human tumors indicate that the FHIT gene is altered by mutations, predominantly deletions, in human cancer and that such alterations are common in epithelial cancers, particularly those resulting from exposure to environmental carcinogens, such as lung, head and neck, esophageal, stomach, pancreatic, and cervical cancer [14-16]. FHIT tumor suppressor gene, the work of the mechanism of apoptosis and cell cycle control is still unknown [17-19]. In their study of the FHIT gene is protected with other types of renal cell tumors, but compare carcinoma FHIT expression, they showed that a high proportion of losses. Laryngeal cancer represents one of the most common head and neck malignancies, accounting approximately for 20% of all cases. Many genetic and environmental factors in the development of tumors are known to play a role [40-42] In some tumors, particularly those associated with exposure to environmental carcinogens, alterations in FHIT occur quite early in the development of human cancer. Thus, frequent molecular abnormalities in head and neck squamous cell carcinoma include alterations in tumor suppressor genes p16INK4A, p53, p14ARF, FHIT, RASSF1A, Rb, cyclin D1, and activation of oncogenes, such as members of the ras gene family, c-myc, and EGFR [10,40,29]. FHIT Fragile Histidine Triad gene is associated highly methylated in HNSCC [33] and associated with poor prognosis in early stage esophageal squamous cell carcinoma [41-42]. A few studies has been showed that FHIT expression was associated with locoregional recurrence paradoxically in head and neck squamous cell carcinoma (HNSCC) [10,36]. Mineta et al. showed that low FHIT expression correlated with high Ki-67 expression, suggestion that neoplastic cells with low FHIT expression may have high proliferatiion potential [40]. Tai et al. found that patients classified as high risk who that a FHIT negative tumor experienced locoregional recurrence less often (%18) that did high risk patients who that a FHIT pozitive tumor(%33) [36]. Otero et al. implicated that overexpression of 53p predicted a trend toward an improved prognosis, where as no reduction in FHIT expression predicted a significantly poorer outcome in patients with advanced oropharyngeal cancer [20]. We believe that further investigation into FHIT gene expression will be helpful in clarifying the biological mechanisms of larynx carcinogenesis as well as in the early detection and prevention of larynx cancer. But in addition to this findings, the present findings highlight that the FHIT gene partially related to target for analysis in case with laryngeal squamous cell carcinoma. According to results of analyses, we idendified a link in between the exon 8 and exon 9 of FHIT gene. However, studies with larger number of patients will be needed to establish this finding. These experiments will indicate whether the complete absence of FHIT protein results in the development of a spectrum of tumors in the mouse, as might be expected from the study of the genetic alterations at the FHIT locus in human tumors. If this could be achieved, it could result not only in better treatment of FHIT negative tumors but also in the ability to eliminate or reduce precancerous lesions, contributing to the prevention of some of the most common human cancers.
We are grateful to Dr. Şahande Elagöz for assistance in obtaining the clinical material used in this study and to Dr. Hasibe Cingilli VURAL for her support of this research program.
- Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM (2002) FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol 3: 748-754. [crossref]
- Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, et al. (1996) The FHIT gene 3p14.2 is abnormal in lung cancer. Cell 85: 17-26. [crossref]
- Scully C, Field JK, Tanzawa H (2000) Genetic aberrations in oral or head and neck squamous cell carcinoma 2: chromosomal aberrations. Oral Oncol 36: 311-327. [crossref]
- Thiagalingam S, Lisitsyn NA, Hamaguchi M, Wigler MH, Willson JK, et al. (1996) Evaluation of the FHIT gene in colorectal cancers. Cancer Res 56: 2936-2939. [crossref]
- Zekri AR, Bahnassy AA, Hafez M, El-Shehaby AM, Sherif GM, et al. (2005) Alterations of the fragile histidine triad gene in hepatitis C virus-associated hepatocellular carcinoma. J Gastroenterol Hepatol 20: 87-94. [crossref]
- Yoo HJ, Yun BR, Kwon JH, et al. (2009) Genetic and expression alterations in association with the sarcomatous change of cholangiocarcinoma cells. Experimental and molecular medicine 41: 102-115. [crossref]
- Myers NE, Suen JY. (1996) Cancer of the head and surgery. Philadelphia: WB Saunders; p. 381-2.
- Dores GM, Anderson WF, Curtis RE, Landgren O, Ostroumova E, et al. (2007) Chronic lymphocytic leukaemia and small lymphocytic lymphoma: overview of the descriptive epidemiology. Br J Haematol 139: 809-819. [crossref]
- Paveli AK, Krizanac S, Cacev T, Hadzija MP, Radosevi AS, et al. (2001) Aberration of FHIT gene is associated with increased tumor proliferation and decreased apoptosis-clinical evidence in lung and head and neck carcinomas. Mol Med 7: 442-453. [crossref]
- Otero-Garcia JE, Youssef E, Enamorado II, Du W, Yoo GH, et al. (2004) Prognostic significance of p53 and FHIT in advanced oropharyngeal carcinoma. Am J Otolaryngol 25: 231-239. [crossref]
- Panagopoulos I, Thelin S, Mertens F, Mitelman F, Aman P (1997) Variable FHIT transcripts in non-neoplastic tissues. Genes Chromosomes Cancer 19: 215-219. [crossref]
- Latil A, Bièche I, Fournier G, Cussenot O, Pesche S, et al. (1998) Molecular analysis of the FHIT gene in human prostate cancer. Oncogene 16: 1863-1868. [crossref]
- Guo Z, Johansson SL, Rhim JS, Vishwanatha JK (2000) Fragile histidine triad gene expression in primary prostate cancer and in an in vitro model. Prostate 43: 101-110. [crossref]
- Rohr UP, Rehfeld N, Geddert H, Pflugfelder L2021 Copyright OAT. All rights reservelevance of fragile histidine triad protein expression in patients with small cell lung cancer. Clin Cancer Res 11: 180-185. [crossref]
- Burke L, Khan MA, Freedman AN, Gemma A, Rusin M, et al. (1998) Allelic deletion analysis of the FHIT gene predicts poor survival in non-small cell lung cancer. Cancer Res 58: 2533-2536. [crossref]
- Guler G, Uner A, Guler N, Han SY, Iliopoulos D, et al. (2004) The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer 100: 1605-1614. [crossref]
- Semba S, Huebner K (2006) Protein expression profiling identifies cyclophilin A as a molecular target in Fhit-mediated tumor suppression. Mol Cancer Res 4: 529-538. [crossref]
- Fouts RL, Sandusky GE, Zhang S, Eckert GJ, Koch MO, et al. (2003) Down-regulation of fragile histidine triad expression in prostate carcinoma. Cancer 97: 1447-1452. [crossref]
- Eyzaguirre EJ, Mietttinen M, Norris BA, et al., (1999) Different immunohistochemical patterns of Fhit protein expression in renal neoplasm. Molecular Pathology 12: 979-983.
- Dumon KR, Ishii H, Fong LY, Zanesi N, Fidanza V, et al. (2001) FHIT gene therapy prevents tumor development in Fhit-deficient mice. Proc Natl Acad Sci U S A 98: 3346-3351. [crossref]
- Boring CC, Squires TS, Tong T, Montgomery S (1994) Cancer statistics, 1994. CA Cancer J Clin 44: 7-26. [crossref]
- Karakök M, Aydin A, Erkiliç S, et al., (2001) Epidemiological evaluation of cancers diagnosed in Gaziantep State and SSK Hospitals laboratories. Pathology Bulletin 18: 10-15.
- Perez-Ordoñez B, Beauchemin M, Jordan RC (2006) Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol 59: 445-453. [crossref]
- Partridge M, Emilion G, Pateromichelakis S, Phillips E, Langdon J (1999) Location of candidate tumour suppressor gene loci at chromosomes 3p, 8p and 9p for oral squamous cell carcinomas. Int J Cancer 83: 318-325. [crossref]
- Hogg RP, Honorio S, Martinez A, et al. (2002) Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 321 in head and neck squamous cell carcinoma. Eur J Cancer 38: 1585-1592.
- Inal E, Kemaloglu Y, Bayramoglu I, et al. (1993) Retrospective analysis of our cases with laryngeal cancer. K.B.B. And Bass Neck Surgery Magazine 1: 23-28.
- Çankaya H, Garça MF, Miçoogullari ED, et al. (2013) Approaching Laryngeal Cancer Patients. Van Tip Dergisi 20: 8-12
- Williams HK (2000) Molecular pathogenesis of oral squamous carcinoma. Mol Pathol 53: 165-172. [crossref]
- Forastiere A, Koch W, Trotti A, Sidransky D (2001) Head and neck cancer. N Engl J Med 345: 1890-1900. [crossref]
- Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, et al. (2004) Cancer statistics, 2004. CA Cancer J Clin 54: 8-29. [crossref]
- el-Naggar AK, Hurr K, Batsakis JG, Luna MA, Goepfert H, et al. (1995) Sequential loss of heterozygosity at microsatellite motifs in preinvasive and invasive head and neck squamous carcinoma. Cancer Res 55: 2656-2659. [crossref]
- Ambatipudi S, Gerstung M, Gowda R, Pai P, Borges AM, et al. (2011) Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS One 6: e17250. [crossref]
- Kim MM, Califano JA (2004) Molecular pathology of head-and-neck cancer. Int J Cancer 112: 545-553. [crossref]
- Kim YT, Park JY, Jeon YK, et al. (2009) Aberrant promoter CpG island hypermethylation of the adenomatosis polyposis coli gene can serve as a good prognostic factor by affecting lymph 210 Methylation - From DNA, RNA and Histones to Diseases and Treatment node metastasis in squamous cell carcinoma of the esophagus. Dis Esophagus 22:143-1450.
- Lee EJ, Lee BB, Kim JW, Shim YM, Hoseok I, et al. (2006) Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma. Eur J Cancer 42: 972-980. [crossref]
- Tai SK, Lee JI, Ang KK, El-Naggar AK, Hassan KA, et al. (2004) Loss of Fhit expression in head and neck squamous cell carcinoma and its potential clinical implication. Clin Cancer Res 10: 5554-5557. [crossref]
- Ragin CC, Taioli E (2007) Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer 121: 1813-1820. [crossref]
- Tutar E, Karatas E, Baglam T. (2010) Expression of the Fragile Histidine Triad Gene in Laryngeal Carcinoma. International Journal of Hematology and Oncology 20: 229-234.
- Ahmad A, Ansari MA, Mudassar S, et al. (2009) Promoter Hypermethylation in Tumor Suppressing Genes p16 and FHIT and Their Relationship with Estrogen Receptor and Progesterone Receptor Status in Breast Cancer Patients from Northern India. Translational Oncology 2: 264-270. [crossref]
- Mineta H, Miura K, Takebayashi S, Misawa K, Ueda Y, et al. (2003) Low expression of fragile histidine triad gene correlates with high proliferation in head and neck squamous cell carcinoma. Oral Oncol 39: 56-63. [crossref]
- Lee JI, Soria JC, Hassan K, Liu D, Tang X, et al. (2001) Loss of Fhit expression is a predictor of poor outcome in tongue cancer. Cancer Res 61: 837-841. [crossref]
- Mutlu C, Unal A, Altunbas A, et al. (1996) Retrospective analysis of our patients with total laryngectomy. SDU Tip Faculty magazine 3: 21-25.








