MicroRNAs (miRNAs) are small non-coding RNAs which play important roles in cellular process including response to viral infection. When cell is infected with virus, the cellular miRNAs expression may be changed and affect both host and viral gene expression. Some viruses can also produce miRNAs which can target directly to both host and viral mRNAs. Nowadays IFNs have been widely used as an anti-tumor therapeutics agents due to anti-proliferative actions. IFNs signaling pathways promote IFN-inducible genes including microRNAs (miRNAs). They can induce the target cells to become anti-viral phase rapidly by triggering the IFN signaling pathways acting as anti-viral activators. In this review, we summarized the role of miRNAs in virus-host interaction including effects of miRNAs to interferon which is widely used for viral infection treatment.
interferon, microRNA, virus-host interaction
Interferons are parts of non-specific immune system (also called innate immune system) which play an important role to defense against viral infections by induction of proteins involved in anti-viral defenses and stimulated the host immune response [1]. Interferons are produced by cells that responded to viral nucleic acids and rapidly induced target cells into anti-viral phase at early stage of viral infection. Inactive formed of IFNs are not created in order to accumulate in cells (Figure 1) [2]. IFNs can be classified into three classes [3], depending on IFNs specific receptors show in Figure 2. First, type I IFNs, which can be produced by virus-infected cells and leukocytes. Members of type I IFNs included IFN α (more than 14 subtypes), β, δ, ε, κ, and ω which can be recognized by type I IFN receptor complex (IFNAR) [4,5]. Some of type I interferons have been reported that they are found in human, therefore IFN- α, IFN- β, IFN-ε, IFN-κ, IFN-ω, and then triggered IFN signaling pathways including signal transducers and activators of transcription (STATs) and Janus kinases (JAKs) to promote IFN-stimulated genes (ISGs) acting as anti-viral activity. Previous studies reported that some proteins can stimulate IFNs induction such as retinoic-acid-inducible protein I (Rig-I) helicase and melanoma-differentiation-associated gene 5 (MDA5) which can recognize the viral RNA in cytoplasm of virus-infected cells [6]. Second, type II IFNs, which can be produced by activated T lymphocytes and natural killer cells (NK cells), are also called “immune interferon”. This class includes IFN-γ which preferentially binds to IFNγR ( also called IFNGR),captivating to IFNGR1 and IFNGR2 activated through JAK-STATs pathway, following by the production of a Gamma Activation Factor (GAF), one of the cytoplasmic factor in latent phase, which later preferentially binds to the Gamma Activation Site (GAS) on promotor region [7]. Lastly, type III IFNs, which consist of IFN-λ binding to IFNλ -specific receptor (IFNλR). IFN I receptors are predominantly specific to epithelial-originated cells. The dominant biological activities and expression of type III IFN are almost identical to type I IFNs [1].
Figure 1. Diagram demonstrate the mechanism of viral infection can be inhibited by the stimulation of Interferon Stimulate Gene (ISG) and alternated miRNAs expression pattern.
Figure 2.IFN-signaling pathway, three classes of IFNs Types I, II and III binding to specific IFN receptor. IFNs Types I, III receptor activates same signaling pathways by phosphorylate STAT1 and 2 by JAK1 and Tyrosine kinases 2 (TRK2) results STAT proteins form heterodimers implicate with IRF9 bind to IFN-stimulated response elements (ISREs) activates associate genes transcription. In Type II IFNs activates by phosphorylate STAT1 by JAK1 and JAK2 results STAT proteins form homodimers bind to IFN gramma-activated site (GAS)
At present, IFNs have been now developed and used widely and continuously in pharmaceuticals and clinical interventions. There are some differences of indications of applying interferons in medical specialties. Type I IFNs, is found to be badly absorbed from digestive tract. Therefore, they are usually applied in other manner besides digestive tract; e.g. intravenous or intramuscular injection. Moreover, this type of IFNs showed a short time circulation in administration due to the long periods of low drug levels following by the sporadic anti-viral effects. However, the solution for pitfalls of IFNs usage has been developed by the increase proportion of using bio-pharmaceuticals products such as enzymes, peptides, proteins, oligonucleotides and antibody in pharmaceutical market worldwide [8].
Although there have been reported more than 14 subtypes of IFN-α until present, especially IFN-α2 has been used in therapeutic methods. Many types of novel poly-material have been used to conjugate with IFN-α to increase its medical efficiencies, such as polyethylene glycol (PEG) and albumin protein, which is also called recombinant IFNs. The propose of conjugation is to increase solubility and also decrease proteolysis, antigenicity and clearance in renal pathway [9,10]. Some commercial products of PEG-conjugated IFN-α products have been merchandised in market, for example, PegIntron® (peginterferon α-2b) and Pegasys® (peginterferon α-2a). However, these trade names of interferons showed the various forms of PEG and interferons themselves. Therefore, the pegylated IFNα2a generated from using the combination of 9 isomers of branched PEG (40 kDa in weight of each isomer) while peginterferon α-2b generated from 14 isomers of linear PEG (12 kDa each), conjugated with IFN-α-2b. The medical treatment of interferon α has been widely effectively used against chronic HBV and HCV infection, which can lead to chronic Hepatitis symptoms [11,12]. At present, the IFN-β therapy is known to be as the initial treatment or also called the first-line therapy for relapsing-remitting multiple sclerosis and whist. The utilizing of IFN-β can bring about the concerns owing to the neutralizing effects of antibodies against them.
Peter et al., 2014 show primary results from the first 48 weeks (placebo-controlled period) of the 2-year, phase 3 [13], The efficacy and safety of using PEG-interferon β-1a has been evaluated from recent study in the application of PEG-IFNβ1a for 2 weeks or 4 weeks in relapsing-remitting multiple sclerosis patients. The study showed that after recombinant PEG-IFNβ1a has been treated for 48 weeks, the deterioration rate could reduce significantly compared with placebo. This study suggested that the PEG-conjugated drug can be resulted in longer period of treatments than normal frequent administration for relapsing-remitting multiple sclerosis.
For type II IFNs, the human IFN-gamma gene, situated on chromosome 12, contains three introns and four exons coded for a polypeptide of 166 amino acids in length, 20 of which constitute the signal peptide. Previous research suggested that the IFN-γ treatment was secured and effectual in patients with chronic granulomatous disease. Moreover, the progression of IFN-γ prophylaxis in chronic granulomatous disease is resembled to be better and have excellent toleration over the time sustainment [14]. The treatment of IFN-γ can improve the anti-fungal immune level while the extended studies have guaranteed to evaluate the effectiveness of clinical outcome of IFN-γ treatment. Recombinant Human Interferon-gamma (IFN-γ), a bioactive protein has been used in cell culture applications, can be manufactured in E.coli culture in a single, non-glycosylated, polypeptide chain. Previous study also refer that the recombinant IFN-γ (rIFN-γ) treatment showed the constructive effects on the fungal infection consequences in patients with chronic granulomatous disease [15]. Current study shows that the pegylated IFN-λ, one of type III interferons, is in phase III clinical trials of chronic HCV therapy [16]. Potentially, IFN-λ might be one of effective therapeutic agents against both HCV and HBV infection. Preliminary clinical studies have appeared that IFN-λ1 may exhibit comparable anti-viral efficiency as type I IFN with less side effects [17].
MicroRNAs (miRNAs) are the highly conserved groups of small non-coding RNAs which play a critical role in term of endogenous specific gene silencing through the RNA interference (RNAi) pathway in several multicellular organisms. Over past decades, many evidences supported that the miRNAs have been regulated the gene expression by the translational repression or mRNA degradation. To date, over 1,000 regions of human genome (approximately 3%) have been discovered to encode miRNAs. Moreover, the studies suggested that around 40% of miRNA-coding genes are lied down in intron regions or non-coding exon regions of the human genome [18].
The biogenesis of miRNAs started from the transcription of miRNA-coding genes called miR-gene in nucleus by RNA polymerase II [19,20]. After the transcription, the RNA folded to form multiple hairpin-loops with 5¢ capping and 3¢ poly A tail called primary microRNA or pri-miRNA [19,21], followed by the cropping process via the microprocessing complex composed of; an RNase III endonuclease called Drosha; and the important cofactor with RNA recognizing domain called Digeorge Syndrome critical region 8 (DGCR8) to generate the precursor microRNA (pre-miRNA) with approximately 60-70 nucleotides in length [22]. After that, the pre-miRNAs would be exported to cytoplasm by Exportin-5 (XPO5) mediated transport using energy from Ran-GTP [23]. In the cytoplasmic process, the pre-miRNAs would be cleaved by the RNase III enzyme called Dicer [24] to generate the mature miRNA duplex with approximately 21-23 nucleotides in length consisted of the guide (antisense) strand and the passenger (sense) strand. Then the mature miRNA duplex would be incorporated into the RNA-induced silencing complex (RISC) composed of several proteins essentially argonaute-2 (AGO2), GEMIN3 and GEMIN4. After incorporation into RISC, the miRNA duplex unwinding would be facilitated by helicase activity of the GEMIN3 to discard the passenger strand of miRNA out of the complex whereas the guide strand of miRNA with RISC were directed to the 3′ untranslated region (UTR) of the specific target messenger RNA via base complementary between miRNA and mRNA [25,26]. The perfect complementary binding between miRNA and its target mRNA triggers mRNA cleavage and degradation by AGO2 [27]. On the other hand, the partial complementary can lead to translational repression (Figure 3) [28].
Normally, the animal miRNAs are able to recognize its targeted sequences on mRNA partially, especially 6-8 nucleotides from 5' direction, which is called seed region [29,30]. The effective base complementary between miRNA and mRNA can be classified into 3 patterns including 5'-seed, 5'-canonical and 3'-compensatory. The 5'-seed contains significant base pairing within seed region (position 2nd-8th from 5'-end of miRNA) while 5'-canonical employs complementary in seed region and additional base pairing in 3'-end of miRNA. The 3'-compensatory yields base complementary at least half of the sequence within 3'-end of miRNAs without any base-pairing in seed region (Figure 3) [31].
Figure 3. Biogenesis of miRNAs, miRNA-genes are transcribed into primary miRNAs by RNA polymerase II (Pol II) after they were cleaved by Dosha and DGCR8 to pre-miRNAs. The pre-miRNAs are exported to cytoplasm by Expotin5 (XPO5) and further processed by Dicer to become miRNA duplex, which incorporated into the RNA-induced silencing complex (RISC). These molecules complementarily bind to target mRNAs, leading to mRNA degradation or translation repression.
Several of cellular mechanisms can be regulated by miRNAs, such as cell proliferation signaling, chromosome maintenance, cell differentiation and cell apoptosis [25,32-35]. Mainly, miRNAs are endogenous, but some studies revealed the extracellular effects of miRNAs in cell-cell signal transduction, which can be used as a reasonable description that sometimes the miRNAs can be evaluated in blood serum, plasma or saliva. The previous study suggested that more than 50 species of miRNAs can be harvested from saliva of healthy people and some can be changed significantly due to the disease progression, for example, miR-200a and miR-125a have been found particularly in the saliva of oral cancer patients [36].Thus, these miRNAs in body fluids can be used as the potential biological markers in clinical diagnosis.
Combine with the further knowledge that miRNAs, which can complementary bind to various target genes, can be regulated by IFN. Here in, we hypothesized that the side effects from IFN treatment in patient might result from the downstream effects of gene regulation process via miRNA mechanisms.
The interferon activities have been involved in main part of cellular innate immune system. Suggesting that the endogenous IFN-regulated miRNAs are also associated in the anti-viral defense mechanism after virus infection while the targeted host genes are also essential for IFN activation using the regulation of virus-encoded miRNAs after antigen recognition [1].
Interferon induces host miRNAs expressions
The up-to-date high-throughput sequencing is advantageous to identify miRNA content expressed in human samples in stable stage, compared to endogenous miRNAs expressed during specific conditions, such as viral infection. The results demonstrated the differential expression of miRNAs between 2 groups of cells under conditions, comparable with distinctive IFNs levels, suggesting that IFNs are critical for regulation of some IFN-inducible miRNAs mechanisms (Table 1).
Table 1. IFNs regulated microRNA
| Type of IFNs |
IFNs |
miRNAs |
Regulation |
Target of miRNA |
References |
| I |
IFN α |
miR-122 |
↓ |
3´ UTR HBV mRNA
CyclinG1
HO-1
5´ UTR HCV mRNA |
[39
[116]
[117] |
| miR-130a/301 |
↑ |
3´ UTR of SMAD4 |
[40] |
| MiR-203 |
↑ |
IFIT1/ISG56 |
[41] |
| IFN β |
miR-155 |
↑ |
SOCS1 |
[118]
[119] |
| miR-29a |
↑ |
HIV genome 3´ UTR |
[46] |
| miR-122 |
↓ |
3´ UTR HBV mRNA
CyclinG1
HO-1
5´ UTR HCV mRNA |
[39]
[117]
[37] |
| miR-21 |
↓ |
PDCD4 |
[42] |
| miR-26a
miR-34a
Let-7b |
↑ |
PTEN
Ezh2 histone methyltransferase,
p53 |
[44] |
| II |
IFNγ |
miR-29a |
↑ |
GAS-elements |
[120] |
| miR-155 |
↑ |
SOCS1 |
[49] |
| miR-520b |
↑ |
MICA |
[50] |
| III |
IFNλ |
miR-15a |
↑ |
Bcl-2 |
[51] |
Abbreviations. Bcl-2: B-cell CLL/lymphoma 2; EZH2: Enhancer of zeste homolog 2; HMT: histone methyltransferase; GAS: Gamma interferon activation site; HO-1: Heme Oxygenase 1; HBV: Hepatitis B virus; HCV: Hepatitis C virus; HIV: Human immunodeficiency virus; IFIT1/ISG56: Interferon induced protein with tetratricopeptide repeats 1; MICA: MHC class I-related chain A; PTEN: Phosphatase and tensin homolog; PDCD4: Programmed cell death 4; SMAD4: SMAD family member 4; SOCS1: Suppressor of cytokine signaling 1; p53: Tumor protein p53
One of interesting Type I IFN-stimulating miRNAs, miR-122, which facilitates the hepatitis C virus (HCV) replication processes. Previous study revealed that miR-122 can be reduced due to IFN signal in order to confine HCV replication [37,38]. Besides, previous studies reported the existing of miR-122 binding element in the 3′UTR of mRNAs of all 4 genes in Hepatitis B virus therefore, the pre-C/C or pre-genomic RNA (pgRNA), pre-S, S, and X mRNAs [39]. Previous study revealed the possibility of anti-tumor response of IFN-α by the treatment of HCV-infected Huh7.5 cells with IFN-alpha could up-regulates miR-130a/301 while it caused c-Met expression reduction and HCV pathogenesis [40].
An IFN-inducible miRNA, miR-203 was found to negatively trigger a bunch of cellular mRNAs, inclusive of an IFN-stimulated gene target, IFIT1/ISG56, by diminishing the stabilization of its mRNA transcript [41]. Previous study expressed that the IFN-β can inhibit the expression of miR-21 via the experiment of the addition of STAT3 activator in order to induce the IFN-β resulted in miR-21 suppression. In contrast, the STAT3 inhibitor vector can terminate the suppression regulation of miR-21 [42], which plays the crucial role as the in malignant gliomas [43]. Treatment of IFN-β in primary macrophages could result in the up-regulation of the miR-26a, -34a, and let-7b expression, suggesting a negative feedback mechanism in the mediation of IFN- β protein [44,45].
Type I and II IFN that found to be regulated by the same types of miRNA, mir-29 and mir-155. Another IFN-inducible miRNA, miR-29a, has been reported to be induced by IFN-β. The inhibition of miR-29a can bring about the increase of HIV replication while mRNA complex of miR-29a-HIV was found to be co-localized with RISC [46]. MiR-155, which can be induce the phosphorylation process of STAT-1 and 3, resulted in the increase of MxA and ISG15 IFN-regulated anti-viral genes and also has a large anti-viral activity against hepatitis B virus, is up-regulated in NK cells that have the high level of IL-12- and IL-18. This miRNA is also found to be abundant of STAT4 in mice inoculated the cytomegalovirus [47] miR-155 is revealed to regulate SOCS1 gene [48,49] .
For the regulation mechanism of Type II IFNs, it was found that the miR-520b was activated by IFN-γ, resulting in MHC class I-related chain A (MICA) reduction, which refers to surface protein levels. Interestingly, miR-520b can perform on both the MICA 3'UTR and also as the promoter region. Moreover it affected a reduction in the levels of MICA transcript that NKG2D ligand associated with the down-regulation of NKG2D ligand MICA expression [50].
The study found the correlation in up-regulation between type III IFN and plasma autoantibodies and plasma miR-15a levels in lupus mouse model. The up-regulation of miR-15a may suggest the promotion of cellular miRNAs packaging for translocation to extracellular fluids including plasma [51]. The miR-15a boosting can case the apoptosis due to the anti-apoptotic protein bcl-2, one of target gene of miR-15a [52].
MiRNAs regulate IFNs response
Knowing to be the first barrier of innate immune response, IFNs have been proved that they can be triggered by miRNAs and vice versa. In many previous studies suggested that the expression of miRNAs can be up-regulated or down-regulated by viral infection. Interestingly, the viral genome can also encode to be miRNA, may be to facilitate itself in propagation process [53]. In this review, we stated some examples of relationship between IFNs and miRNA in virus infection mechanisms (Table 2).
Table 2. microRNAs regulated IFNs
| miRNAs |
Regulated IFN-receptors & IFN-R signaling |
References |
| miR-22 |
IRF5 |
[54] |
| miR-9 |
IFI44L, PSMB8, IRF5, PSMB10, IFI27, IFIT2, TRAIL, IFIT1 and IRF1 |
[55] |
| miR-466l |
IFN α |
[57] |
| miR-1231 |
IFNAR1 |
[58] |
| miR-146a |
IFN β |
[56] |
| miR-26a |
IFN β |
[44] |
| miR-34 |
IFN β |
| Let-7b |
IFN β |
| miR-29 |
IFNγ, GAS-elements |
[59]
[60]
[121] |
| miR-155 |
SOS1, IFN-γRα |
[122]
[123] |
| miR-548 |
IFN-λ1 |
[61]
[62] |
Abbreviations: GAS: Gamma interferon activation site; IFN-γRα: IFN-gammaR alpha; IFN α: Interferon alpha; IFNAR1: Interferon alpha and beta receptor subunit 1; IFI27: Interferon alpha inducible protein 27; IFN β: Interferon beta; IFN γ: Interferon gamma; IFI44L: Interferon induced protein 44 like; IFIT1: Interferon induced protein with tetratricopeptide repeats 1; IFIT2: Interferon induced protein with tetratricopeptide repeats 2; IFN-λ1: Interferon lambda 1; IRF1: Interferon regulatory factor 1; IRF5: Interferon regulatory factor 5; IRF5: Interferon regulatory factor 5; PSMB10: Proteasome subunit beta 10; PSMB8: Proteasome subunit beta 8; SOS1: SOS Ras/Rac guanine nucleotide exchange factor 1; TRAIL: Tumor necrosis factor superfamily member 10.
MicroRNAs can also mediate IFNs mechanisms. For example, the expression of interferon gene can be down-regulated by miR-22 when the miR-22 addressed directly to high mobility group box-1 and interferon regulatory factor (IRF)-5, resulting in activation prevention of IRF3 and NF-ĸB, which downstream modulate series of interferon genes [54].
MiR-9 has been found to be significantly associated with immunity and inflammatory diseases regulation. In addition, the miR-9 could trigger the IFN-induced genes expression (e.g., IFI44L, PSMB8, IRF5, PSMB10, IFI27, IFIT2, TRAIL, IFIT1 and IRF1) and MHC class I molecules (e.g., HLA-B, HLA-H, HLA-C and HLA-F) in human cancer cells [55].
MicroRNAs regulated Type I IFNs
Some previous studies discovered the miRNAs regulation in type I interferons, for example, vesicular stomatitis virus (VSV) infection can increase the expression of miR-146a in macrophages. The related pathway in miR-146a is RIG-I/NF-ĸB-dependent pathways which associated with Type I IFN [56]. The previous experiment performed in macrophages and dendritic cells transfected with miR-466l expression, followed by Sendai virus (SeV) and vesicular stomatitis virus (VSV) infection. The results of this study showed the suppression of interferon-alpha (IFN-α) expression, suggesting that the miR-4661 can regulate IFN-α family [57]. The seed sequence of miR-466l is AUAAAUA, and it was found to be complementary to the typical AU-rich elements (ARE) located in the 3′ UTR of multiple cytokines, chemokines and growth factors. miR-466l targets the located in the 3′ UTRs of many IFN- α species, including IFN- α1, - α2, - α4, - α8, - α10, - α13, - α16, - α17 and - α21, thus inhibiting IFN- α production and enhancing viral replication [57].
Human miR-1231 was upregulated in response to HBV infection in human hepatocytes hsa-miR-1231 suppresses HBV replication at the post-transcriptional level but not through the activation of interferon signaling. The previous evidences suggested that miR-1231 can regulate IFNAR1 which associate in type I membrane protein synthesis of IFNα and β receptor [58]. The study found that the hsa-miR-1231 significantly increased in hepatocyte of HBV-infection patients, resulted in the inhibition of HBV propagation in post-transcriptional level, not interferon signaling inhibition.
MicroRNAs regulated Type II IFNs
In infecting with L. monocytogenes, the host cell can be vulnerable to the infection by miR-29 which can repress the production of IFN-γ of NK cells to damage the host immune response [59]. MiR-29 which also regulate IFN-γ production and helper T cell differentiation and can be the biological marker for human diseases that are related to T cell-mediated immunity such as asthma, type 1 diabetes, and multiple sclerosis [60].
MicroRNAs regulated Type III IFNs
The 3′ UTR of IFN- λ1 has been reported as the target of miRNA-548 family, including miR-548b-5p, miR-548c-5p, miR-548i, miR-548j, and miR-548n. Advance study identified that the expression of IFN-λ1 gene was down-regulated by miRNA-548 mimics transfection while the complementary RNAs, acting as inhibitors, can increase the IFN-stimulated genes and IFN- λ1 itself [61]. Human miR-548 family members are significantly engaged with the undermined IFN signaling of CHB. Correspondingly, hsa-miR-548ah-5p expression is obviously developed in the immune provocation phase of CHB. Host anti-viral response might be down-regulated via direct targeting of IFN-λ1 [62].
Perspective of IFNs
IFNs have been widely used as the anti-tumor therapeutics agents due to the anti-proliferative actions. They also have been used as anti-viral agents during both of acute and chronic viral infection diseases since they can induce the target cells to become anti-viral phase rapidly by triggering the IFN signaling pathways acting as anti-viral activators. Many studies has been shown the effective clinical benefits of mixing IFNs in cancer and viral infection trials, either in monotherapies or using IFNs in combination with other therapeutics agents, e.g. chemotherapeutics drugs. Some defects of using interferons treatment have been improved by using recombinant interferons such as PEG-conjugated interferons and other biopharmaceuticals products to increase medical efficiency. However, using IFNs treatment still brings about some side effects in patients which we hypothesized that it might result from the downstream effects of gene regulation processes, one of which is associated with miRNAs regulations. Previous studies have been discovered that IFNs can be triggered by miRNAs and vice versa. The endogenous miRNAs which can regulate a bunch of genes in many cellular processes can be triggered by the variation of IFNs level. The change of mRNAs level due to miRNAs suppression or activation can cause the reversal of cellular mechanisms and leads to side effects which sometimes these genes are not the direct targets of IFNs treatment at all, which should be considered before subjecting of IFN treatment.
In addition, miRNAs play roles in virus-host interaction. Cellular miRNAs expression levels are changed when cell is infected with virus. These miRNAs may target both host and viral genes similar to viral miRNAs which can be expression by some viruses also regulate both host and viral genes (Figure4). When host mRNAs are targeted by host miRNAs or viral miRNAs, the translation of these genes will be repressed. While the regulation of viral mRNAs by both host miRNAs or viral miRNAs can support or inhibit viral replication.
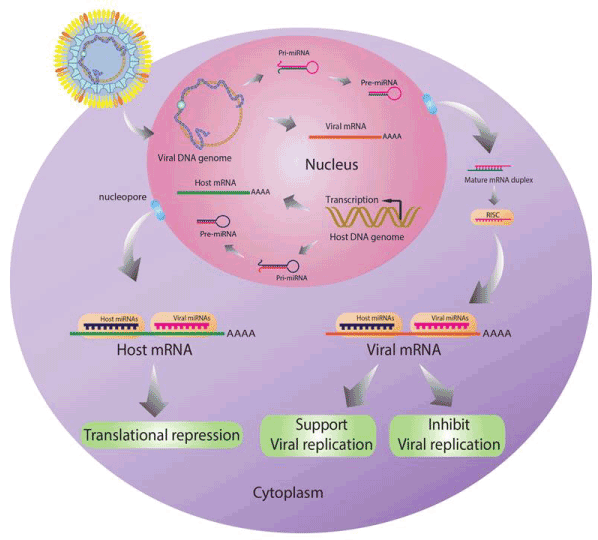
Figure 4. Roles of miRNAs in virus-host interaction. Host miRNAs can inhibit both cellular and viral gene translation. While the viral miRNAs also target both host and viral genes. The regulation of these miRNAs may support or inhibit viral replication.
Many host miRNAs can inhibit viral replication by targeting viral genomes or mRNAs and silencing their expression (Table 3). For example, hsa-miR-1231 which showed high homology with core and HBx sequences of hepatitis B virus (HBV). The hsa-miR-1231 can suppress HBV replication by HBV core reduction [63]. Hsa-miR-24 and hsa-miR-638 were also reported to be candidate anti-viral host-encoded miRNAs inhibiting HBV replication. These miRNAs decreased the levels of HBV transcripts or HBV gene products in HepG2 cell culture model [64] . Hsa-miR-125a was also reported to inhibit HBV replication by targeting the viral transcript encoding particle surface antigen. Besides, the expression of the hsa-miR-125a was increased in both HBV patients [65]. Cellular hsa-miR-141 was found to be repressed HBV expression and replication. The hsa-miRNA-141 was reported to regulated host peroxisome proliferator-activated receptor alpha (PPARA) leading to repress the HBV promoter activities [66].
Table 3. Host miRNAs inhibit viral replication
| Viruses |
Host miRNAs |
Targeted viral genes |
Effect to viral replication |
References |
| HBV |
miR-1231 |
core and HBx |
suppress |
[63] |
miR-24
miR-638 |
- |
suppress |
[64] |
| miR-125a |
surface antigen |
suppress |
[65] |
| PFV-1 |
miR-32 |
- |
suppress |
[71] |
| IAV |
miR-323
miR-491
miR-654 |
PB1 |
suppress |
[68] |
| miR-3145 |
PB1 |
suppress |
[69] |
| let-7c |
M1 |
suppress |
[70] |
| HTLV-1 |
miR-28-3p |
gag/pol mRNA |
suppress |
[72] |
| DENV |
let-7c |
- |
suppress |
[73] |
| VSV |
miR-24 |
L protein |
suppress |
[74] |
| miR-93 |
P protein |
suppress |
| HIV-1 |
miR-28
miR-125b
miR-150
miR-223
miR-382 |
- |
suppress |
[75] |
| miR-29a |
Nef |
suppress |
[76] |
| miR-132 |
MeCP2 |
promote |
[77] |
| HCV |
miR-122 |
S1 and S2 |
promote |
[78] |
| let-7b |
NS5B |
suppress |
[80] |
| HCMV |
miR-200 family |
UL122 |
promote |
[79] |
| KSHV |
miR-1258 |
RTA |
suppress |
[81] |
miR-498
miR-320d |
RTA |
suppress |
[82] |
| EV71 |
miR-296-5p |
Capsid VP3 & VP1 |
suppress |
[83] |
| miR-23b |
EV71 3' UTR |
suppress |
[84] |
Abbreviations: HBV: Hepatitis B virus ; PFV-1: Primate foamy virus type 1; IAV: Influenza A virus; HTLV-1: Human T-Cell Lymphotropic Virus type 1; DENV: Dengue virus; VSV: Vesicular stomatitis virus; HIV-1: Human immunodeficiency virus type 1; HCV: Hepatitis C virus; HCMV: Human cytomegalovirus; KSHV: Kaposi's sarcoma-associated herpesvirus; EV71: Enterovirus 71; HBx: Hepatitis B virus X gene; PB1: Polymerase basic 1; Nef: Negative regulatory factor ; MeCP2: Methyl-CpG binding protein 2; NS5B: Nonstructural protein 5B; RTA: Replication and transcription activator
In addition, some studies suggested computational method to predict host miRNAs targeting to viral genomes i.e. hsa-miR-489, hsa-miR-325, hsa-miR-876-3p and hsa-miR-2117 were predicted to target HA, PB2, MP and NS of influenza H1N1 virus, leading to the inhibition of viral replication [67] while miRNA hsa-miR-323, hsa-miR-491 and hsa-miR-654 target PB1 gene of H1N1 influenza A virus (A/WSN/33). The repression of PB1 expression, the component of viral polymerase complex leads to inhibition of influenza A virus replication [68]. The hsa-miR-3145 was also predicted to target PB1 gene of influenza A viruses subtype pH1N1, H5N1 and H3N2. The luciferase assay confirmed that the miR-3145 directly targeting PB1 gene of influenza A viruses. In addition, the in vitro transfection of miR-3145 expression vector confirmed that the miR-3145 can inhibit viral replication [69]. Human let-7c was reported to be upregulated in influenza virus-infected A549 cells. Nucleotide sequence on seed region of let-7c is perfect complementary to the 3′ UTR of viral M1 gene. In vitro study confirmed that let-7c can down-regulate the expression of viral M1 at both in the RNA and protein levels [70].
Human miR-32 is host miRNA that can target primate foamy virus type 1 (PFV-1) and reduces the viral accumulation in host cells [71]. Hsa-miR-28-3p is highly expressed in resting T cells. These cells can resist to human T cell leukemia virus, type 1 (HTLV-1) infection. This miRNA targets HTLV-1 mRNA specifically in genomic gag/pol site and represses viral replication. Moreover, the polymorphism in hsa-miR-28-3p target site in ATK-1 strain which cannot be inhibited by hsa-miR-28 resulting in its efficiently transmission [72] .
Overexpression of human let-7c can inhibit dengue viruses (DENV) both DENV-2 and DENV-4 replication in human hepatoma Huh-7 cells. The study of Escalera-Cueto et al. also reported that BACH1 which targeted by let-7c was downregulated due to DENV infection [73]. The repression of transcription factor BACH1 led to the upregulation of heme oxygenase 1 (HO-1) which causes the stress responding in infected cells [73].
For Vesicular stomatitis virus (VSV), hsa-miR-24 was reported to target viral large protein (L protein) and hsa-miR-93 also targeted to phosphoprotein (P protein) encoded by VSV. The repression of these viral genes suggested that cellular hsa-miR-24 and hsa-miR-93 can inhibit VSV replication [74].
Cellular hsa-miR-28, hsa-miR-125b, hsa-miR-150, hsa-miR-223 and hsa-miR-382 target 3' end of human immunodeficiency virus type 1 (HIV-1) transcripts and repress viral genes expression. In activating CD4+ T cells, this group of miRNAs were downregulated, allowing HIV to replicate in these cells [75]. Furthermore, the luciferase assay confirmed that HIV-1 Negative factor (Nef) gene contains the target site of cellular hsa-miR-29a. The suppression of viral Nef gene by this miRNA results in the inhibiting of viral replication [76]. The miR-132 is upregulated in CD4+ T cell activation due to HIV-1 infection. This miRNA targets MeCP2 and regulates its expression. In contrast to other miRNAs, the function of the miR-132 was found to be enhanced HIV-1 replication [77].
Further host miRNAs can inhibit viral replication; some host miRNAs also promote viral replication. For example, hsa-miR-122 targets to 5′ UTR of S1 and S2 genes of hepatitis C virus (HCV) and promotes its life cycle. It also can support viral replication by stabilizing viral RNAs and protecting them from host cytoplasmic exoribonuclease, Xrn1 [78]. Moreover, hsa-miR-200 miRNA family was suggested to promote human cytomegalovirus (HCMV) latency by targeting the viral UL122 genes [79].
Human let-7b was also suggested to suppress the HCV replication. Let-7b targets 5′ UTR of NS5B and regulates its expression leading to inhibit viral replication. In addition, the target site of let-7b in NS5B are conserved among various HCV genotypes [80].
Negative factor (Nef) is a secreted HIV-1 protein upregulating cellular miRNAs including miR-1258. The luciferase assay was performed and confirmed that miR-1258 directly targeted a seed sequence in the 3′ UTR of the replication and transcription activator (RTA), the major lytic switch protein which controls Kaposi’s sarcoma-associated herpesvirus (KSHV) reactivation. The upregulation of miR-1258 reduced RTA synthesis and inhibited KSHV replication. This finding suggested that miR-1258 may inhibit KSHV replication to promote viral latency in pathogenesis of AIDS-related malignancies [81]. Herpes simplex virus (HSV)-1 is an important cofactor in KSHV reactivation by inducing the expression of KSHV replication and transcription activator (RTA). The HSV-1 can downregulate cellular miR-498 and miR-320d. These two host miRNAs target the 3′ UTR of KSHV RTA. Overexpression of these miRNAs inhibited HSV-1-induced KSHV replication. In addition, miR-498 or miR-320d without HSV-1 infection also regulates KSHV by targeting RTA [82].
The expression of hsa-miR-296-5p was significantly increased in enterovirus 71 (EV71)-infected cells. The overexpression of hsa-miR-296-5p inhibits EV71 replication by targeting to EV71 genome (nt 2115 to 2135 and nt 2896 to 2920 in BrCr strain) located in the EV71 genome. These target sites were validated by luciferase reporter assays and Western blotting. However, the mutated target sequences of the EV71 HeN strain did not show the inhibitory effects of the miR-296-5p. Thus, the miR-296-5p can inhibit virus infection and that the virus mutates to escape suppression by cellular miRNAs [83]. The hsa-miR-23b is also downregulated in EV71-infected cells. In silico analysis and luciferase assay were performed to validate the target site of miR-23b in viral VP1 gene. In vitro transfection of mimic miR-23b verified its effects on inhibition of EV71 replication. These results suggest that miR-23b and upregulation of miR-23b inhibited the replication of EV71 by targeting at EV71 3′UTR conserved sequence by regulating the expression of viral VPl [84].
Despite host miRNAs can target viral genomes or viral mRNAs, some target host genes and inhibit viral replication (Table 4). Hsa-miR-130a targets PGC1α and PPARγ which are metabolic regulators stimulating HBV replication. In HBV-infected hepatocytes, this miRNA was found to be downregulated. Therefore, the expression of both PGC1α and PPARγ are increased and stimulates the HBV replication [85]). Hsa-miR-26b was suggested to inhibit HBV production including antigen expression, transcription, and replication. In vitro study showed that miR-26b significantly repressed HBV enhancer/promoter activities. For host gene, miR-26b targets the cysteine- and histidine-rich domain containing 1 (CHORDC1) which increased viral activities by promoting HBV enhancer/promoter activities. During HBV infection, the miR-26b is suppressed and the expression of CHORDC1 is increased. In addition, the miR-26a has a similar anti-HBV function as miR-26b [86]. Cellular miR-501 is up-regulated in HepG2 cells with HBV replication. The expression of miR-501 expression is also up-regulated in hepatocellular carcinoma tissues. Inhibition of miR-501 can inhibit HBV replication. An inhibitor of HBV replication, HBXIP is the target of miR-501. The regulation of HBXIP by miR-501 induces HBV replication [87].
Table 4. Host miRNAs targets host genes and effect to viral replication
| Viruses |
Host miRNAs |
Targeted host genes |
Effect to viral replication |
References |
| HBV |
miR-130a |
PGC1α and PPARγ |
suppress |
[85] |
| miR-26b |
CHORDC1 |
suppress
| [86] |
| miR-501 |
HBXIP |
promote |
[87] |
| miR-141 |
PPARA |
suppress |
[66] |
| miR-29c |
TNFAIP3 |
suppress |
[88] |
| HCV |
miR-199a-5p |
PI3K/Akt, Ras/ERK, Wnt/β-catenin |
promote |
[89] |
| HIV-1 |
let-7c, miR-34a, miR-124a |
P21, TASK1 |
promote |
[90] |
| miR-155, miR-181a |
SAMHD1 |
promote |
[91] |
| JEV |
miR-146a |
TRAF6, IRAK1, IRAK2, STAT1 |
promote |
[92] |
| CHIKV |
miR-146a |
TRAF6, IRAK1 and IRAK2 |
promote |
[93] |
| DENV |
let-7c |
BACH1 |
suppress |
[73] |
| miR-30e-3p |
IκBα |
suppress |
[94] |
| HCMV |
miR-21 |
Cdc25a |
suppress |
[95] |
| EV71 |
miR-27a |
EGFR |
suppress |
[96] |
| IAV |
miR-4276 |
COX6C |
suppress |
[97] |
| miR-29c |
A20 mRNA protection |
promote |
[98] |
| CBV3 |
miR-126 |
SPRED1 |
promote |
[99] |
| miR-203 |
ZFP-148 |
promote |
[100] |
| SIV |
miR-29a, miR-29b, miR-9, miR-146a |
Nef/U3 , R region |
suppress |
[101] |
| HSV-1 |
miR-101 |
ATP5B |
suppress |
[102] |
Abbreviations: HBV: Hepatitis B virus; HCV: Hepatitis C virus; HIV-1: Human immunodeficiency virus type 1; JEV: Japanese encephalitis virus; CHIKV: Chikungunya virus; DENV: Dengue virus; HCMV: Human cytomegalovirus; EV71: Enterovirus 71 ; IAV: Influenza A virus; CBV3: Coxsackievirus B3; SIV: Simian immunodeficiency virus; HSV-1: Herpes simplex virus type 1; PGC1α: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; PPARγ: Peroxisome proliferator-activated receptor gamma; CHORDC1: Cysteine And Histidine-Rich Domain Containing 1, HBXIP: Hepatitis B virus X-interacting protein; PPARA: Peroxisome proliferator-activated receptor alpha; Tumor Necrosis Factor, TNFAIP3: Alpha-Induced Protein 3 ; PI3K/Akt: Phosphoinositide 3-kinase/Protein kinase B ; Ras/ERK: Ras/Extracellular signal-regulated kinase ; SAMHD1: SAM domain and HD domain-containing protein 1; TRAF6: TNF receptor associated factor 6; IRAK1: Interleukin-1 receptor-associated kinase 1; IRAK2: Interleukin-1 receptor-associated kinase 2; STAT1: Signal transducer and activator of transcription 1; BACH1: Transcription regulator protein BACH1; IκBα: Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha; Cdc25a: Cell division cycle 25 homolog A; EGFR: Epidermal growth factor receptor; COX6C: Cytochrome c oxidase subunit 6C; SPRED1: Sprouty-related, EVH1 domain-containing protein 1; ZFP-148: Zinc finger protein 148; ATP5B: ATP synthase subunit beta, mitochondrial
The cellular miR-29c is significantly downregulated in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) cells. It targets tumor necrosis factor alpha-induced protein 3 (TNFAIP3) which plays roles in inflammation and immunity. Overexpression of miR-29c in HepG2.2.15 cells significantly repressed the expression of TNFAIP3 and leaded to the inhibition of cell proliferation, induction of apoptosis and HBV DNA replication [88].
Human miR-199a-5p was upregulated in hepatitis C virus (HCV) infected. The miR-199a-5p may promote the replication of HCV by regulating PI3K/Akt, Ras/ERK and Wnt/β-catenin which play important role in pro-survival pathways. Moreover, the inhibition of miR-199a-5p can repress the expression of NS3 and NS5A as well as the HCV RNA. Therefore, the upregulated miR-199a-5p was suggested to support the replication of HCV [89].
The expression of let-7c, miR-34a and miR-124a, which were upregulated due to HIV-1 infection could target and regulate host p21 and TASK1. The inhibition of these genes increasing viral replication in infected cells. In addition this study suggested that HIV-1 may use these host cellular miRNAs to inhibit the innate immune mechanism and allow more viral replication efficiency [90].
Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1) limits the HIV infection. The SAMHD1 was reported to be regulated by cellular miR-155 and miR-181a. Overexpression of these miRNAs repressed the expression of SAMHD1 and also increased HIV-1 replication. In the study of Pilakka-Kanthikeel and colleagues, found that the expression of SAMHD1 was increased in human astrocytes and restricted the HIV replication [91].
Japanese encephalitis virus (JEV) upregulates the expression of cellular miR-146a. This miRNA can regulate TRAF6, IRAK1, IRAK2 and STAT1 genes leading to the suppression of NF-κB activation and discontinuance of anti-viral Jak-STAT pathway. This function of upregulated miR-146a helps JEV to evade the immune response. In addition, this effect of miR-146a was suggested to be strain specific [92]. Similar to JEV, the Chikungunya virus infection upregulates the expression of cellular miR-146a which targets TRAF6, IRAK1 and IRAK2. The regulation of these genes by miR-146a modulates host anti-viral immune and increases replication of CHIKV [93].
Infection of dengue virus (DENV) upregulates the expression of host miRNA let-7c. The overexpression of let-7c can repress DENV-2 and DENV-4 replication. For host gene, let-7c targets transcription factor BACH1 which downregulated during the DENV infection. This regulation of BACH1 suggested that HO-1 the main responsive factor of BACH1 was up-regulated and led to the oxidative stress response in DENV-infected cells [73]. The miR-30e-3p (miR-30e*) was also reported to be upregulated during DENV infection. It directly targets the 3′ UTR of IκBα leading to the hyperactivation of NF-κB. This upregulation of NF-κB increases the expression of downstream IFN-stimulated genes including OAS1, MxA and IFITM1. The increasing of these innate immunity associated genes suppress DENV replication [94].
Infection human cytomegalovirus (HCMV) downregulates the expression of cellular miR-21 and leads to the upregulation of miR-21 targeted gene, Cdc25a which is a cell cycle regulator. Overexpression of miR-21 in vitro inhibited viral activities including gene expression, genome replication and infectious progeny production. The overexpression of Cdc25a promoted viral replication. In addition, three viral genes IE1, pp71 and UL26 were suggested to inhibit the transcription of miR-21. Therefore, the host cellular miR-21 may be an anti-viral factor for HCMV by regulating Cdc25a [95].
In enterovirus 71 (EV71)-infected cells, the expression of miR-27a was revealed to be significantly decreased. While the in vitro over-expression of miR-27a shown to inhibit the replication of EV71. In silico analysis also suggested that miR-27a targeting host EGFR mRNA. Luciferase assay was performed and confirmed that miR-27a could target EGFR and repress its expression. This regulation of EGFR also decreases the Akt and ERK phosphorylation which facilitates EV71 replication. Therefore, the miR-27a can inhibit the replication of EV71 by modulating the expression of EGFR [96].
Influenza virus infection can change host miRNAs expression profile. Cellular miR-4276 was found to be downregulated in the early stage of influenza virus infection. The downregulation of miR-4276 led to the upregulated of cytochrome c oxidase VIC (COX6C), the target gene. COX6c expression also related to the expression of apoptotic protein caspase-9. The miR-4276 may repress viral replication by inducing the COX6C and caspase-9 [97]. The miR-29 family is also upregulated during influenza A virus infection. But only miR-29c was reported to involve in the viral replication. The miR-29c can induce the expression of A20 in infected cells by protecting A20 mRNA from degradation. The upregulation of miR-29c and A20 expression are involved in regulation of the innate immune response which can promote viral replication [98].
MicroRNAs were also reported to regulate coxsackievirus B3 (CVB3) infections. The cellular miR-126 which upregulated during CBV3 infection regulates two signal pathways essential for CVB3 replication. First, the miR-126 suppresses EVH1 domain containing 1 (SPRED1) and enhances ERK1/2 activation promoting CVB3 replication. The miR-126 also stimulates GSK-3β activity and induced degradation of β-catenin through LRP6 and WRCH1 suppression, which informed the cells to virus-induced cell. The regulation of SPRED1, LRP6, and WRCH1 genes by miR-126 promotes CBV3 replication [99]. In addition, the miR-203 is also upregulated during CVB2 infection. It is upregulated through the activation of protein kinase C/transcription factor AP-1 pathway. It targets a transcription factor called zinc finger protein-148 (ZFP-148). The regulation of ZFP-148 translation by miR-203 increases cell viability and enhances CVB3 replication [100].
MiRNAs also regulate the replication of the simian immunodeficiency virus (SIV). In silico analysis predicted that Nef/U3 and R regions of SIV RNA contains binding sites for miR-29a, miR-29b, miR-9 and miR-146a. These four miRNAs decreases viral production and viral RNA expression. In addition, each of these miRNAs was regulated by TNFα and/or the type I IFN, IFNβ [101].
Endogenous hsa-miR-101 can inhibit the replication of herpes simplex virus-1 (HSV-1). The miR-101 targets 3'UTR of mitochondrial ATP synthase subunit beta (ATP5B) and regulates its expression. ATP5B is a pro-viral factor, the regulation of this gene by miR-101 inhibiting HSV-1 replication [102].
MiRNAs can also be transcribed by viruses which reported to target both viral genes and host genes (Table 5). The regulation of viral genes by viral miRNAs may involve in the disease progression such as changing viral cycle into lytic phase. In addition, the regulation of some host genes promotes viral replication, miR-M3 of Marek’s disease virus type 1 targets host SMAD2 and inhibits the apoptotic process in infected cells. Human cytomegalovirus (HCMV) can escape host immune respond by encoding miR-UL112 which targets host MICB. It also encodes miR-US25-1 targeting CCNE2 which may inhibit the cell cycle and prevent apoptosis.
Table 5. Viral miRNAs targets host genes and effect to viral replication
| Viruses |
Viral miRNAs |
Targeted viral/host genes |
Effect to viral replication |
References |
| MDV1 |
miR-M3 |
Host: SMAD2 |
Anti-apoptotic |
[125] |
| miR-M4 |
Host: PU.1 |
Mimic cellular miR-155 |
[112] |
| Host: GPM6B |
immune tolerance |
[113] |
| Host: RREB1 |
immune tolerance |
[113] |
| Host: c-Myb, |
immune tolerance |
[113] |
| Host: MAP3K7IP2 |
- |
[113] |
| Host: C/EBP |
- |
[113] |
| Virus: UL28 and UL32 |
Cleavage/packaging of herpesvirus DNA |
[113] |
| HCMV |
miR-UL112 |
Host: MICB |
Immune escape |
[114] |
| miR-UL112-1 |
Virus: UL114 |
Prevent lytic replicaton/ promote latency |
[124] |
| miR-US25-1 |
Host: CCNE2 |
Block cell cycle to prevent apoptosis |
[115] |
Abbreviations: MDV1: Marek's disease virus serotype 1; HCMV: Human cytomegalovirus; SMAD2: Mothers against decapentaplegic homolog 2; PU.1: Transcription factor PU.1; GPM6B: Neuronal membrane glycoprotein M6-b; RREB1: Ras-responsive element-binding protein 1; c-Myb : Myb proto-oncogene protein; MAP3K7IP2: Mitogen-activated protein kinase kinase kinase 7-interacting protein 2; C/EBP: CCAAT-enhancer-binding proteins; MICB: MHC class I polypeptide-related sequence B; CCNE2: Cyclin E2
The hcmv-miR-US25-1-5p is encoded from HCMV. It is highly expressed during lytic and latent phase of infections. It is also suggested to inhibit viral replication by targeting viral YWHAE, UBB, NPM1, and HSP90AA1 genes. Luciferase assay and western blot analysis were performed to confirm the regulation of these genes by hcmv-miR-US25-1-5p [103]. In addition, human herpesvirus 6A (HHV-6A), member of herpesvirus family encodes viral miRNA, miR-U86. This miRNA targets the HHV-6A gene, U86 which play essential role in viral life cycle [104]. The HCMV also encodes hcmv-miR-US33 which can inhibit the lytic viral replication. It also targets host Syntaxin3 (STX3) gene which had been validated by Hybrid-PCR and luciferase-reporter assays. In miR-US33-5p overexpression cells downregulated the expression of STX3 protein, leading to inhibition of HCMV DNA synthesis and viral replication [105]. The miR-UL70-3p which encoded from HCMV was reported to target host MOAP1 and PHAP while miR-UL148D was also suggested to target host ERN1 which plays a role in the mitochondrial-dependent intrinsic pathway of apoptosis. The regulation of these pro-apoptotic genes by HCMV miRNAs showed that HCMV uses its miRNAs to inhibit the cellular apoptosis which support viral replication in infected cells [106] .
The miR-US25-2-3p encoded from HCV reduces viral replication by regulating eukaryotic translation initiation factor 4A1 (eIF4A1). In vitro study confirmed that miR-US25-2-3p decreased HCMV and host genomic DNA synthesis. It also inhibited host cell proliferation. However, the transfection of miR-US25-2-3p inhibitor upregulated eIF4A1 and increased HCMV copy number. Thus, the over-expression of miR-US25-2-3p decreases eIF4A1 expression and contributes to the inhibition of HCMV replication [107].
The miR-BART20-5p is one of the viral miRNAs encoded from Esptein-Barr virus (EBV). This miRNA targets two EBV genes, BZLF1 and BRLF1. The BRLF1 3′ UTR contains two seed match sites which reported to be target sites of the miR-BART20-5p. The transfection of miR-BART20-5p mimic demonstrated the suppression of various EBV early proteins and the production of viral progeny also. Thus, miR-BART20-5p plays an important role in EBV latency maintenance [108]. The miR-BART18-5p is encoded from the BamH1 fragment A rightward transcript (BART) region of EBV. This miRNA target the 3′ UTR of MAP3K2 which is the same site as cellular miR-26a-5p. The regulation of MAP3K2 inhibits EBV lytic viral replication and maintains latency phase [109].
BK polyomavirus (BKV) encodes a viral miRNA, miR-B1 which upregulated during viral infection. In vitro transfection of miR-B1 expression vector demonstrated that inhibition of Tag expression led to the repression of Tag-enhanced promoter activity and viral replication. The miR-B1 was also suggested to be used in a potential treatment strategy against BKV infection [110].
As the result from deep sequencing, the miR-H3 was reported to be viral miRNA encoded from HIV-1. The miR-H3 is encoded from viral mRNA region encoding the active center of reverse transcriptase (RT) which are conserved among various subtypes of HIV-1 viruses. The miR-H3 promotes viral replication by upregulating HIV-1 RNA transcription and protein expression. The miR-H3 interacts with TATA box in HIV-1 5' LTR and upregulates the promoter activity activating HIV-1 latency [111].
Herpesvirus-encoded miR-K12-11 was recently shown to be a functional ortholog of miR-155, a miRNA that plays a major role in lymphoid malignancies and the modulation of immune responses. Here we show that miR-M4, encoded by the highly oncogenic Marek's disease virus of chickens, shares common targets with miR-155 and thus is also a functional ortholog of miR-155, the first one identified in an alphaherpesvirus. The observation showed that two distinct oncogenic herpesviruses associated with distinct types of lymphomas in different species encode functional miR-155 orthologs suggested the importance of this miRNA in regulatory pathways and the biology of lymphomagenesis [112].
Mdv1-miR-M4 is one of 25 miRNAs expressed by Marek's disease virus (MDV-1. Mdv1-miR-M4 was shown to be the second functional viral ortholog of miR-155, a cellular miRNA that plays a crucial role in several physiological and pathological processes in lymphocyte biology. Using luciferase reporter assays, we showed that mdv1-miR-M4-5P and miR-155 efficiently targeted a common set of 3' untranslated regions (3' UTR) of six cellular genes (GPM6B, RREB1, c-Myb, MAP3K7IP2, PU.1 and C/EBP) [112,113]. In addition, we also investigated the interactions between mdv1-miR-M4-5P and mdv1-miR-M43P and viral mRNAs encoding UL28 and UL32 in both reporter and western blot assays. Mdv1-miR-M4 specifically inhibited the translation of these two viral proteins, which are involved in the cleavage/packaging of herpesvirus DNA [113].
Human cytomegalovirus miRNAs, HCMV-miR-UL112 was reported to target the major histocompatibility complex class I-related chain B (MICB) gene. MICB is a stress-induced ligand of the natural killer (NK) cell activating receptor NKG2D and is critical for the NK cell killing of virus-infected cells and tumor cells. We showed that hcmv-miR-UL112 specifically down-regulates MICB expression during viral infection, leading to decreased binding of NKG2D and reduced killing by NK cells. This reveals a miRNA-based immunoevasion mechanism that appears to be exploited by human cytomegalovirus [114].
The miR-UL112 of HCMV was also reported to downregulate the expression of a host immune gene, MICB. Remarkably, it was shown that the same miRNA also downregulates immediate-early viral genes and that its ectopic expression resulted in reduced viral replication and viral titers. The targets for most of the viral miRNAs, and hence their functions, are still unknown. The miR-UL112 also targets the UL114 gene, and the reduction of UL114 by miR-UL112 reduces its activity as uracil DNA glycosylase but only minimally affects viral growth. In addition, two additional HCMV-encoded miRNAs, miR-US25-1 and miR-US25-2 reduce the viral replication and DNA synthesis not only of HCMV but also of other viruses, suggesting that these two miRNAs target cellular genes that are essential for viral growth. Thus, we suggest that in addition to miR-UL112, two additional HCMV miRNAs can control the life cycle of the virus [114].
The miR-US25-1 binds target sites primarily within 5′ UTRs, mediating significant reduction in gene expression. Intriguingly, many of the genes targeted by miR-US25-1 are associated with cell cycle control, including cyclin E2, BRCC3, EID1, MAPRE2, and CD147, suggesting that miR-US25-1 is targeting genes within a related pathway. Deletion of miR-US25-1 from HCMV results in over expression of cyclin E2 in the context of viral infection. Our studies demonstrate that a viral miRNA mediates translational repression of multiple cellular genes by targeting mRNA 5′ UTRs [115].
In conclusion, when viruses infect human cells, many of cellular processes respond to the infection including IFN activities and expression of cellular miRNAs. Due to the function of miRNAs which can regulate host genes expression via mRNAs degradation or translational repression, cells may use these small non-coding RNAs to regulate responded genes. The effects of miRNAs can support or alter IFN activities, directly inhibit viral replication or stimulate host immune system. In the other hand, many cellular miRNAs may induce apoptosis, support viral replication or increase viral production. Responded miRNAs are determined by specificities of virus and host cell types. Roles of miRNAs may be clues for systematic explanation of virus-host interactions.
Funding was supported by a Joint Research Program between Thailand and Japan (NRCT-JSPS); the Thailand Research Fund (TRF: RSA5680031); the Research Chair Grant, the National Science and Technology Development Agency (NSTDA); Chulalongkorn Academic Advancement into Its 2nd Century Project; the Development and Promotion of Science and Technology Talents Project (DPST); the Scholarship from the Graduate School, Chulalongkorn University to commemorate 72nd Anniversary of his Majesty King Bhumibol Adulyadej and the 90th Anniversary Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund).
The authors hereby declare no personal or professional conflicts of interest with any aspect of this review.
- David M (2010) Interferons and microRNAs. J Interferon Cytokine Res 30: 825-828. [Crossref]
- Jensen S, Thomsen AR (2012) Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86: 2900-2910. [Crossref]
- Katsoulidis E, Kaur S, Platanias LC (2010) Deregulation of Interferon Signaling in Malignant Cells. Pharmaceuticals 3: 406-418.
- Fensterl V, Sen GC (2009) Interferons and viral infections. Biofactors 35: 14-20. [Crossref]
- Allen G, Fantes KH (1980) A family of structural genes for human lymphoblastoid (leukocyte-type) interferon. Nature 287: 408-411. [Crossref]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101-105. [Crossref]
- Shuai K, Schindler C, Prezioso VR, Darnell JE Jr (1992) Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258: 1808-1812. [Crossref]
- Kang JS, Deluca PP, Lee KC (2009) Emerging PEGylated drugs. Expert Opin Emerg Drugs 14: 363-380. [Crossref]
- Hamidi M, Azadi A, Rafiei P (2006) Pharmacokinetic consequences of pegylation. Drug Deliv 13: 399-409. [Crossref]
- Harris JM, Chess RB (2003) Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov 2: 214-221. [Crossref]
- Santantonio TA, Fasano M (2014) Chronic hepatitis B: Advances in treatment. World J Hepatol 6: 284-292. [Crossref]
- Khaliq S, Latief N, Jahan S (2014) Role of different regions of the hepatitis C virus genome in the therapeutic response to interferon-based treatment. Arch Virol 159: 1-15. [Crossref]
- Calabresi PA, Kieseier BC, Arnold DL, Balcer LJ, Boyko A, et al. (2014) Pegylated interferon beta-1a for relapsing-remitting multiple sclerosis (ADVANCE): a randomised, phase 3, double-blind study. Lancet Neurol 13: 657-65. [Crossref]
- Marciano BE, Wesley R, De Carlo ES, Anderson VL, Barnhart LA, et al. (2004) Long-term interferon-gamma therapy for patients with chronic granulomatous disease. Clin Infect Dis 39: 692-699. [Crossref]
- Delsing CE, Gresnigt MS, Leentjens J, Preijers F, Frager FA, et al. (2014) Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: a case series. BMC Infect Dis 14: 166. [Crossref]
- Miller DM, Klucher KM, Freeman JA, Hausman DF, Fontana D, et al. (2009) Interferon lambda as a potential new therapeutic for hepatitis C. Ann N Y Acad Sci 1182: 80-87. [Crossref]
- Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, et al. (2010) Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology 52: 822-832. [Crossref]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902-1910. [Crossref]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051-4060. [Crossref]
- Zhou X, Ruan J, Wang G, Zhang W (2007) Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3: e37. [Crossref]
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957-1966. [Crossref]
- Gregory RI, Chendrimada TP, Shiekhattar R (2006) MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 342: 33-47. [Crossref]
- Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185-191. [Crossref]
2021 Copyright OAT. All rights reserv
- Lund E, Dahlberg JE (2006) Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol 71: 59-66. [Crossref]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [Crossref]
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350-355. [Crossref]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437-1441. [Crossref]
- Chu CY, Rana TM (2006) Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210. [Crossref]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787-798. [Crossref]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15-20. [Crossref]
- Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3: e85. [Crossref]
- Loveday EK, Svinti V, Diederich S, Pasick J, Jean F (2012) Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J Virol 86: 6109-22. [Crossref]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999-3004. [Crossref]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. (2004) Human MicroRNA targets. PLoS Biol 2: e363. [Crossref]
- Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, et al. (2011a) miR-200a Prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes 60: 280-7. [Crossref]
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, et al. (2009) Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 15: 5473-5477. [Crossref]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, et al. (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449: 919-922. [Crossref]
- Filipowicz M (2011) Prognostic potential of hepatic miR-122 measurements and antisense strategies targeting miR-122 as a therapeutic approach in viral hepatitis. Liver Int 3: 437-9. [Crossref]
- Li C, Wang Y, Wang S, Wu B, Hao J, et al. (2013) Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol 87: 2193-205.
- Zhang X, Daucher M, Armistead D, Russell R, Kottilil S (2013b) MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-alpha. PLoS One, 8: e55733. [Crossref]
- Buggele WA, Horvath CM (2013) MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56. J Virol 87: 9260-9270. [Crossref]
- Ohno M, Natsume A, Kondo Y, Iwamizu H, Motomura K, et al. (2009) The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res 7: 2022-2030. [Crossref]
- Chen Y, Liu W, Chao T, Zhang Y, Yan X, et al. (2008) MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 272: 197-205. [Crossref]
- Witwer KW, Sisk JM, Gama L, Clements JE (2010) MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol 184: 2369-2376. [Crossref]
- Seitz H (2009) Redefining microRNA targets. Curr Biol 19: 870-873. [Crossref]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, et al. (2009) Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696-709. [Crossref]
- Su C, Hou Z, Zhang C, Tian Z, Zhang J (2011) Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J 8: 354. [Crossref]
- Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D (2013) Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155. Proc Natl Acad Sci U S A 110: 6967-72. [Crossref]
- Kim JH, Jou I, Joe EH (2014) Suppression of miR-155 Expression in IFN-γ-Treated Astrocytes and Microglia by DJ-1: A Possible Mechanism for Maintaining SOCS1 Expression. Exp Neurobiol 23: 148-154. [Crossref]
- Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD (2009) Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol 182: 39-43. [Crossref]
- Yuan Y, Kasar S, Underbayev C, Vollenweider D, Salerno E, et al. (2012) Role of microRNA-15a in autoantibody production in interferon-augmented murine model of lupus. Mol Immunol 52: 61-70. [Crossref]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 102: 13944-13949. [Crossref]
- Gottwein E, Cullen BR (2008) Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3: 375-387. [Crossref]
- Polioudakis D, Bhinge AA, Killion PJ, Lee BK, Abell NS, et al. (2013) A Myc-microRNA network promotes exit from quiescence by suppressing the interferon response and cell-cycle arrest genes. Nucleic Acids Res 41: 2239-2254. [Crossref]
- Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, et al. (2013) miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 43: 610-6. [Crossref]
- Hou J, Wang P, Lin L, Liu X, Ma F, et al. (2009) MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK, and IRAK2. J Immunol 183: 2150-2158. [Crossref]
- Li Y, Fan X, He X, Sun H, Zou Z, et al. (2012) MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell Mol Immunol 9: 497-502. [Crossref]
- Zhou C, Yu Q, Chen L, Wang J, Zheng S, et al. (2012) A miR-1231 binding site polymorphism in the 3'UTR of IFNAR1 is associated with hepatocellular carcinoma susceptibility. Gene 507: 95-98. [Crossref]
- Ma F, Xu S, Liu X, Zhang Q, Xu X, et al. (2011) The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol 12: 861-869. [Crossref]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, et al. (2011) MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35: 169-181. [Crossref]
- Li Y, Xie J, Xu X, Wang J, Ao F, et al. (2013) MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-λ1. Protein Cell 4: 130-141. [Crossref]
- Xing TJ, Xu HT, Yu WQ, Wang B, Zhang J (2014) MiRNA-548ah, a potential molecule associated with transition from immune tolerance to immune activation of chronic hepatitis B. Int J Mol Sci 15: 14411-14426. [Crossref]
- Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, et al. (2014) Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J Viral Hepat 2: e89-97. [Crossref]
- Kumar M, Sharma Y, Bandi S, Gupta S (2015) Endogenous antiviral microRNAs determine permissiveness for hepatitis B virus replication in cultured human fetal and adult hepatocytes. J Med Virol 87: 1168-83. [Crossref]
- Mosca N, Castiello F, Coppola N, Trotta MC, Sagnelli C, et al. (2014) Functional interplay between hepatitis B virus X protein and human miR-125a in HBV infection. Biochem Biophys Res Commun 449: 141-145. [Crossref]
- Hu W, Wang X, Ding X, Li Y, Zhang X, et al. (2012) MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One 7: e34165. [Crossref]
- Zhang H, Li Z, Li Y, Liu Y, Liu J, et al. (2013) A computational method for predicting regulation of human microRNAs on the influenza virus genome. BMC Syst Biol 7 Suppl 2: S3. [Crossref]
- Song L, Liu H, Gao S, Jiang W, Huang W (2010) Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84: 8849-8860. [Crossref]
- Khongnomnan K, Makkoch J, Poomipak W2, Poovorawan Y3, Payungporn S4 (2015) Human miR-3145 inhibits influenza A viruses replication by targeting and silencing viral PB1 gene. Exp Biol Med (Maywood) 240: 1630-1639. [Crossref]
- Ma YJ, Yang J, Fan XL, Zhao HB, Hu W, et al. (2012) Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Mol Med 16: 2539-2546. [Crossref]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, et al. (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308: 557-560. [Crossref]
- Bai XT, Nicot C (2015) miR-28-3p is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J Biol Chem 290: 5381-5390. [Crossref]
- Escalera-Cueto M, Medina-Martínez I, del Angel RM, Berumen-Campos J, Gutiérrez-Escolano AL, et al. (2015) Let-7c overexpression inhibits dengue virus replication in human hepatoma Huh-7 cells. Virus Res 196: 105-12. [Crossref]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, et al. (2007) Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27: 123-134. [Crossref]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, et al. (2007) Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13: 1241-1247. [Crossref]
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, et al. (2008) Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 5: 117. [Crossref]
- Chiang K, Liu H, Rice AP (2013) miR-132 enhances HIV-1 replication. Virology 438: 1-4. [Crossref]
- Thibault PA, Huys A, Amador-Cañizares Y, Gailius JE, Pinel DE, et al. (2015) Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5' Untranslated Region. J Virol 89: 6294-6311. [Crossref]
- O'Connor CM, Vanicek J, Murphy EA (2014) Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J Virol 88: 5524-32. [Crossref]
- Cheng JC, Yeh YJ, Tseng CP, Hsu SD, Chang YL, et al. (2012) Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci 69: 2621-2633. [Crossref]
- Yan Q, Ma X, Shen C, Cao X, Feng N, et al. (2014) Inhibition of Kaposi's sarcoma-associated herpesvirus lytic replication by HIV-1 Nef and cellular microRNA hsa-miR-1258. J Virol 88: 4987-5000. [Crossref]
- Yan Q, Li W, Tang Q, Yao S, Lv Z, et al. (2013) Cellular microRNAs 498 and 320d regulate herpes simplex virus 1 induction of Kaposi's sarcoma-associated herpesvirus lytic replication by targeting RTA. PLoS One 8: e55832. [Crossref]
- Zheng Z, Ke X, Wang M, He S, Li Q, et al. (2013) Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J Virol 87: 5645-5656. [Crossref]
- Wen BP, Dai HJ, Yang YH, Zhuang Y, Sheng R (2013) MicroRNA-23b inhibits enterovirus 71 replication through downregulation of EV71 VPl protein. Intervirology 56: 195-200. [Crossref]
- Huang JY, Chou SF, Lee JW, Chen HL, Chen CM, et al. (2015) MicroRNA-130a can inhibit hepatitis B virus replication via targeting PGC1α and PPARγ. RNA 21: 385-400. [Crossref]
- Zhao F, Xu G, Zhou Y, Wang L, Xie J, et al. (2014) MicroRNA-26b inhibits hepatitis B virus transcription and replication by targeting the host factor CHORDC1 protein. J Biol Chem 289: 35029-35041. [Crossref]
- Jin J, Tang S, Xia L, Du R, Xie H, et al. (2013) MicroRNA-501 promotes HBV replication by targeting HBXIP. Biochem Biophys Res Commun 430: 1228-1233. [Crossref]
- Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, et al. (2011) miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 411: 586-592. [Crossref]
- Wang H, Gao H, Duan S, Song X (2015) Inhibition of microRNA-199a-5p reduces the replication of HCV via regulating the pro-survival pathway. Virus Res 208: 7-12. [Crossref]
- Farberov L, Herzig E, Modai S, Isakov O, Hizi A, et al. (2015) MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. J Cell Sci 128: 1607-1616. [Crossref]
- Pilakka-Kanthikeel S, Raymond A, Atluri VS, Sagar V, Saxena SK, et al. (2015) Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1)-facilitated HIV restriction in astrocytes is regulated by miRNA-181a. J Neuroinflammation 12: 66.
- Sharma N, Verma R, Kumawat KL, Basu A, Singh SK (2015) miR-146a suppresses cellular immune response during Japanese encephalitis virus JaOArS982 strain infection in human microglial cells. J Neuroinflammation 12, 30. [Crossref]
- Selvamani SP, Mishra R, Singh SK (2014) Chikungunya virus exploits miR-146a to regulate NF-kB pathway in human synovial fibroblasts. PLoS One 9: e103624. [Crossref]
- Zhu X, He Z, Hu Y, Wen W, Lin C, et al. (2014) MicroRNA-30e* suppresses dengue virus replication by promoting NF-kB-dependent IFN production. PLoS Negl Trop Dis 8: e3088. [Crossref]
- Fu YR, Liu XJ, Li XJ, Shen ZZ, Yang B, et al. (2015) MicroRNA miR-21 attenuates human cytomegalovirus replication in neural cells by targeting Cdc25a. J Virol 89: 1070-1082. [Crossref]
- Zhang L, Chen X, Shi Y, Zhou B, Du C, et al. (2014) miR-27a suppresses EV71 replication by directly targeting EGFR. Virus Genes 49: 373-382. [Crossref]
- Othumpangat S, Noti JD, Beezhold DH (2014) Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 468-470: 256-64. [Crossref]
- Zhang X, Dong C, Sun X, Li Z, Zhang M, et al. (2014) Induction of the cellular miR-29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochem Biophys Res Commun 450: 755-761. [Crossref]
- Ye X, Hemida MG, Qiu Y, Hanson PJ, Zhang HM, et al. (2013) MiR-126 promotes coxsackievirus replication by mediating cross-talk of ERK1/2 and Wntβ-catenin signal pathways. Cell Mol Life Sci 70: 4631-4644. [Crossref]
- Hemida MG, Ye X, Zhang HM, Hanson PJ, Liu Z, et al. (2013) MicroRNA-203 enhances coxsackievirus B3 replication through targeting zinc finger protein-148. Cell Mol Life Sci 70: 277-291. [Crossref]
- Sisk JM, Witwer KW, Tarwater PM, Clements JE (2013) SIV replication is directly downregulated by four antiviral miRNAs. Retrovirology 10: 95. [Crossref]
- Zheng SQ, Li YX, Zhang Y, Li X, Tang H (2011) MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res 89: 219-226. [Crossref]
- Jiang S, Qi Y, He R, Huang Y, Liu Z, et al. (2015) Human cytomegalovirus microRNA miR-US25-1-5p inhibits viral replication by targeting multiple cellular genes during infection. Gene 570: 108-14. [Crossref]
- Nukui M, Mori Y, Murphy EA (2015) A human herpesvirus 6A-encoded microRNA: role in viral lytic replication. J Virol 89: 2615-2627. [Crossref]
- Guo X, Qi Y, Huang Y, Liu Z, Ma Y, et al. (2015) Human cytomegalovirus miR-US33-5p inhibits viral DNA synthesis and viral replication by down-regulating expression of the host Syntaxin3. FEBS Lett 589: 440-446. [Crossref]
- Babu SG, Pandeya A, Verma N, Shukla N, Kumar RV, et al. (2014) Role of HCMV miR-UL70-3p and miR-UL148D in overcoming the cellular apoptosis. Mol Cell Biochem 393: 89-98. [Crossref]
- Qi M, Qi Y, Ma Y, He R, Ji Y, et al. (2013) Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett 587: 2266-2271. [Crossref]
- Jung YJ, Choi H, Kim H, Lee SK (2014) MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol 88: 9027-9037. [Crossref]
- Qiu J, Thorley-Lawson DA (2014) EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci U S A 111: 11157-11162. [Crossref]
- Tian YC, Li YJ, Chen HC, Wu HH, Weng CH, et al. (2014) Polyomavirus BK-encoded microRNA suppresses autoregulation of viral replication. Biochem Biophys Res Commun 447: 543-9. [Crossref]
- Zhang Y, Fan M, Geng G, Liu B, Huang Z, et al. (2014) A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology 11: 23. [Crossref]
- Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, et al. (2009) A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol 83: 489-492. [Crossref]
- Muylkens B, Coupeau D, Dambrine G, Trapp S, Rasschaert D (2010) Marek's disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch Virol 155: 1823-1837. [Crossref]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, et al. (2007) Host immune system gene targeting by a viral miRNA. Science 317: 376-381. [Crossref]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, et al. (2010) A viral microRNA down-regulates multiple cell cycle genes through mRNA 5'UTRs. PLoS Pathog 6: e1000967. [Crossref]
- Hao J, Jin W, Li X, Wang S, Zhang X, et al. (2013) Inhibition of alpha interferon (IFN-α)-induced microRNA-122 negatively affects the anti-hepatitis B virus efficiency of IFN-α. J Virol 87: 137-147. [Crossref]
- Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL (2007) Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology 133: 1166-1174. [Crossref]
- Reinsbach S, Nazarov PV, Philippidou D, Schmitt M, Wienecke-Baldacchino A (2012) Dynamic regulation of microRNA expression following interferon-gamma-induced gene transcription. RNA Biol 9: 978-89. [Crossref]
- Zhang J, Zhao H, Chen J, Xia B, Jin Y, et al. (2012) Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett 586: 3255-3262. [Crossref]
- Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A (2012) Interferon-gamma-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun Signal 10: 41. [Crossref]
- Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, et al. (2012) miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol 189: 1567-1576. [Crossref]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL (2010) Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur J Immunol 40: 225-231. [Crossref]
- Trotta R, Chen L, Ciarlariello D, Josyula S, Mao C, et al. (2012) miR-155 regulates IFN-γ production in natural killer cells. Blood 119: 3478-3485. [Crossref]
- Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, et al. (2009) Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol 83: 10684-93. [Crossref]
- Xu S, Xue C, Li J, Bi Y, Cao Y (2011) Marek's disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J Virol 85: 276-285. [Crossref]