Abnormal shear stress plays a critical role in development of atherosclerosis via modulating complex signal pathways. Long noncoding RNAs (lncRNAs) function directly as RNAs. However, the differential expression of lncRNAs and their net-work regulating the cellular response to the change of shear stress remains to be unclear. Human umbilical vein endothelial cells (HUVEC) were exposure to lower shear stress (2 dynes/cm2) for 120 minutes. Differential expression (defined as a fold change ≥2.0, P<0.05) of lncRNAs and mRNAs was obtained by microarray and analyzed by Box- and Scatter-plot, Heat Map and Hierarchical Clustering. Gene ontology and pathway analyses were performed thereafter. Of 149 lncRNAs differentially expressed between sheared and normal emdothelial cells, 64 lncRNAs were upregulated and 85 lncRNAs were downregulated. 109 mRNAs were differentially expressed (upregulated and 30 downregulated). We found 84 matched lncRNA-mRNA pairs for 62 differentially expressed lncRNAs and 35 differentially expressed mRNAs. 9 pathways through which low shear stress acted on the regulation of cellular function were identified. According to pathway analysis, two up-stream genes (TLR4 and BDNF) were upregulated. Our study provides the expression profiles of lncRNA and mRNA in sheared cells (after 120 min), and reports that TLR4 and BDNF pathways plays crucial role in low shear stress-induced cellular injury, among 84 paired differentially expressed lncRNA-mRNA.
shear stress, endothelial cell, long non-coading RNA, pathway analysis
lncRNA: long non-coding RNA; EC: endothelial cell; HUVEC: Human umbilical vein endothelial cells; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; TLR 4: toll-like receptor 4; LSS: low shear stress
Vascular endothelial cells (ECs) form the inner lining of blood vessel wall, are directly exposed to blood flow pattern, and serve important homeostatic functions in response to various chemical and mechanical stimuli [1,2]. Steady laminar flow (10–20 dyn/cm2) is mandatory to maintain ECs functional by releasing many athero-protective cytokines [3]. In contrast, lower (<10 dyn/cm2) or disturbed blood flow is athero-prone evidenced as atherosclerosis more frequently seen in the inner curvature and bifurcated area of coronary artery tree [4].
Previous studies have reported the differentially expressed genes when Ecs were exposed to lower shear stress or disturbed flow [5-7], with subsequently activation of oxidation, apoptosis, cell death and pro-inflammatory pathways [8,9]. However, the mechanisms regulating those activated genes induced by low shear stress still remains understudied.
Recently, genome-wide analysis have unmasked the extensive transcription of large RNA transcripts that do not code for proteins, termed long noncoding RNAs (lncRNAs), and provide an important new perspective on the centrality of RNA in gene regulation [10]. These lncRNAs function directly as RNAs [11], recruiting chromatin modifiers to mediate transcriptional changes in processes ranging from X-inactivation to imprinting. Unfortunately, there is a lack of information about differential expression of lncRNAs and their net-work regulating the cellular response to the change of shear stress. Accordingly, the current study aimed to provide the genome-wide analysis data to show the differentially expression of lnc RNAs/mRNA and the pathways by which low shear stress acts on the endothelial cells.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVEC) were cultured at 37°C in a humidified incubator with 5% CO2 and maintained in RPMI-1640 culture medium supplemented with 10% fetal bovine serum. When grown to 90% confluence, cells were trypsinized, harvested, resuspended and seeded to a 0.1% gelatin-coated glass. After adherence, monolayer cells grown on the glass were imposed with LSS for different time lengths.
Flow study
A parallel-plate flow chamber that exerts continuous flow was made by sandwiching a silicon gasket between two stainless steel plates with a cover slip sink in the base plate. The chamber and all parts of the circuit tubes were sterilize before the placement of a glass (50 mm × 30 mm) with monolayer cells into the flow chamber. The parameter settings were as follows: flow chamber height, 0.56 mm and pump rate, 60 times/min. The value of shear stress was adjusted by modulating the flow of after-loading and was automatically calculated using information from the pressure transducers. The shear stress in our flow study was 2 dynes/cm2 (for 120 min., marked by ABCD 120, EFGH 120 and IJKL 120) and the cyclic fluid used was 1640 medium with 2% fetal bovine serum supplied. Cultured HUVECs without shear stress were considered as control group (marked by ABCD 0, EFGH 0 and IJKL 0 in Figure 1).
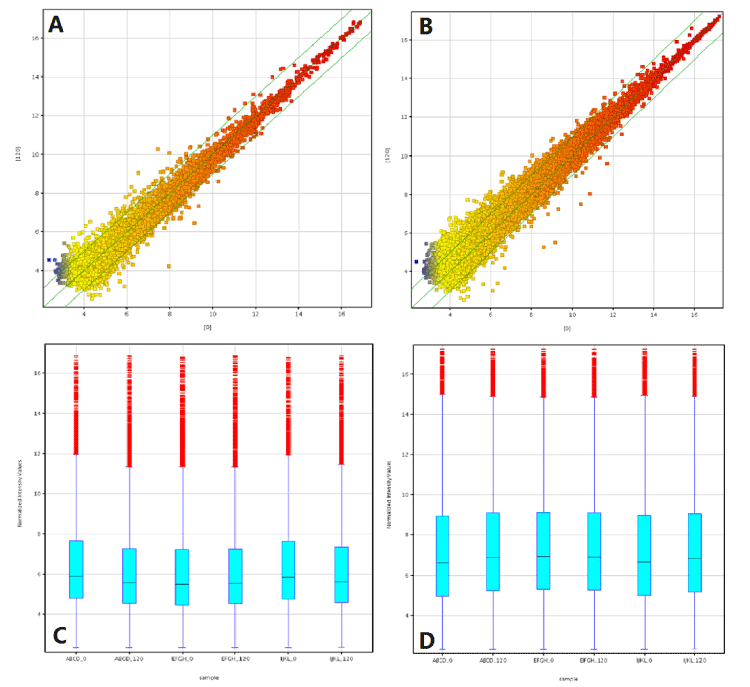
Figure 1. Differentially expressed lncRNA and mRNA in 3 low shear stress-treated vs. paired non-low shear stress treaed endothelial cells. The scatter plot is a visualization method used to assess the lncRNA (A) and mRNA (B) expression variation between paired two group endothelial cells. The value of X- and Y-axes in the scatter plot are the averaged normalized signal values of the group (log2 scaled). The green lines represent fold-change lines (the default fold-change is 2.0). The box plot is a convenient way to quickly visualize the distribution of a dataset of lncRNA (C) and mRNA (D) profiles. After normalization, the distribution of log2 ratios among nine samples were nearly the same.
RNA extraction
Total cellular RNA was isolated from the cells using an RNeasy Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol, and then quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The RNA integrity of each sample was assessed using standard denaturing agarose gel electrophoresis after RNA extraction and prior to sample labeling.
Labeling and hybridization
Total RNA (200 ng each) from cells was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and was reversely transcribed into cDNA using an RNA Spike In Kit with one-color (Agilent) in the presence of 0.8 µL of Random Primer and 2 µL of Spike Mix without 3’ bias utilizing a random priming method. These cDNA samples were then cleaned and labeled in accordance with the Agilent Gene Expression Analysis protocol using Agilent Quick-Amp Labeling Kit. These labeled cDNA samples were used as probes to hybridize to microarrays for 17 h at 65°C using an Agilent Gene Expression Hybridization Kit in Agilent’s SureHyb Hybridization Chambers. The labeled cRNAs were hybridized onto the Human LncRNA Array v3.0 (8 x 60K, Arraystar). After hybridization, the microarrays were washed with an Agilent Wash Buffer kit (Agilent) and scanned with an Agilent microarray scanner G2505C.
lncRNA and mRNA microarray
Arraystar Human LncRNA Microarray V3.0 (Kangcheng Co., Shanghai, China) was designed for the global profiling of human LncRNAs and protein-coding transcripts, with 30,586 LncRNAs and 26,109 coding transcripts being detected. The LncRNAs were carefully constructed using the most highly-respected public transcriptome databases (Refseq, UCSC known genes, Gencode, etc), as well as landmark publications. Each transcript was represented by a specific exon or splice junction probe which can identify individual transcript accurately. Positive probes for housekeeping genes and negative probes were also printed onto the array for hybridization quality control. Each transcript was represented by using 1–5 probes to improve statistical confidence.
Data analysis
The microarrays were scanned at 5 µm/pixel resolutions using an Agilent microarray scanner piloted by GenePix Pro 6.0 software (Axon). Scanned images (TIFF format) were then imported into Agilent Feature Extraction software for grid alignment and expression data analysis. Expression data were normalized by a quantile normalization and the Robust Multichip Average (RMA) algorithm that was included in the Agilent software. Probe-level files and mRNA-level files were generated after normalization. All gene-level files were imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. After quantile normalization of the raw data, lncRNA or mRNAs that at least 3 out of 6 samples have flags in Present or Marginal were chosen for differentially expressed lncRNA or mRNAs screening. Differentially expressed lncRNAs and mRNAs were identified through fold change filtering.
Scatter plot
The Scatter-Plot is a visualization method used for assessing the LncRNA/mRNA expression variation (or reproducibility) between the two compared samples or groups. The values of X and Y axes in the Scatter-Plot were the normalized signal values of the samples or groups (log2 scaled). The green lines were Fold Change Lines (The default fold change value given is 2.0). The LncRNAs/mRNAs above the top green line and below the bottom green line indicated more than 2.0 fold change of LncRNAs/mRNAs between the two compared samples or groups.
Box-Plot
The Box Plot is a convenient way to quickly visualize the distributions of a dataset. It is commonly used for comparing the distributions of the intensities from all samples. After normalization, the distributions of log2-ratios among all tested samples were assessed by this Box Plot.
Differentially expressed LncRNAs screening
To identify significant differentially expressed LncRNAs/mRNAs with statistical significance, we performed a Volcano Plot filtering between the two compared groups. The threshold was Fold Change ≥2.0 and p-value<0.05. Differentially expressed lncRNAs were defined as p value<0.005, FDR<15% and fold-change>2.0.
Heat map and hierarchical clustering
Hierarchical Clustering is one of the most widely used clustering methods for analyzing LncRNA/mRNAs expression data. Cluster analysis arranged samples into groups based on their expression levels, which allowed us to hypothesize the relationships among samples. The dendrogram showed the relationships among LncRNA/mRNA expression patterns of samples.
Gene ontology (GO) analysis
GO analysis is a functional analysis associating differentially expressed genes with GO categories, including biological process, cellular components and molecular function. The GO categories are derived from Gene Ontology (http://www.geneontology.org), which comprise three structured networks of defined terms that describe gene product attributes. The P-value denotes the significance of GO Term enrichment in the differentially expressed gene list. The less the P-value is, the more significant the GO Term is (P-value <= 0.05 was recommended).
Pathway analysis
The predicted target genes above were input into the Database for Annotation. Base on the latest KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg) database, we performed pathway analysis for differentially expressed mRNAs. This analysis allowed to determine the biological pathway that there is a significant enrichment of differentially expressed genes. Furthermore, we also used the Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) and BioCarta (http://www.biocarta.com) to analyze the potential functions of these target genes in the pathways. The P-value denotes the significance of the Pathway. The less the P-value is, the more significant the Pathway is (The P-value cut-off is 0.05).
Statistical analyses
All data were expressed as mean ± standard deviation. Statistical analysis was performed using Student's t-test to compare two variables of microarray data. For example, the statistical significance of a microarray result was analyzed by fold change, and a difference with P<0.05 was considered statistically significant. The false discovery rate was also calculated to correct the P value. The threshold value we used to screen differentially expressed lncRNAs and mRNAs was a fold change ≥2.0 (P<0.05). Furthermore, a differential expression of each lncRNA between sheared and the non-sheared endothelial cells was analyzed using Student's t-test with SPSS (Version 16.0 SPSS, Chicago, IL, USA). P<0.05 was considered significant.
Differentially expressed lncRNAs in endothelial cells
To profile differentially expressed lncRNAs in endothelial cells, we performed a genome-wide analysis of lncRNA and mRNA expression in endothelial cells after treated by low shear stress for 120 min and normal endothelial cells (Figure 1). Using the authoritative data sources, we found that 149 lncRNAs were differentially expressed (fold change ≥2.0, P<0.05) between sheared and normal emdothelial cells. Among them, 64 lncRNAs were upregulated (more than two-fold in sheared vs. the normal cells) and 85 lncRNAs were downregulated (more than two-fold; P<0.05; Table S1). Furthermore, we performed a step-wise genome analysis according to p value. At p value<0.01(more than two-fold), 29 lncRNA were upregulated, with 33 downregulated. When strict criterion (p value<0.005) was used, only 5 lncRNA were upregulated and 8 lncRNA were downregulated (more than two-fold). Finally, when our strict criteria (p value<0.005, FDR<15% and fold-change>2.0) were used, upregulated lncRNAs were C17orf76-AS1, BC079832, and LEF1-AS1; downregulated lncRNA included XLOC-007914 and RP11-473M20.16 (Table S1).
Differentially expressed mRNAs in endothelial cells
In a similar way, we found that 109 mRNAs were differentially expressed (more than two-fold, p<0.05) between sheared and normal endothelial cells (Table S2). Of them, 79 mRNAs were upregulated and 30 downregulated (p<0.05). When p value was designed as less 0.01, 9 mRNAs were upregulated and 7 downregulated, respectively. Finally, by our stric criterion (p<0.005), 6 mRNAs were upregulated and 5 downregulated. We found 84 matched lncRNA-mRNA pairs for 62 differentially expressed lncRNAs and 35 differentially expressed mRNAs (Table S3).
GO and analyses of net-work
Through GO analysis we found that these under-regulated and upregulated transcripts of lncRNAs were associated with cellular process (ontology: biological process), cell (ontology: cellular component), and binding (ontology: molecular function) (Figure 2 and Table S4). Pathway analysis determined that these lncRNAs may target 23 gene pathways that corresponded to transcripts, i.e., “Intracellular Parts” and “Intracellular” composed of 23 targeted genes (with the recommended P-value<0.05) (Figure 2 and Table S5).
Based on these GO data, we then performed the analysis of net-work of differentially expressed lncRNA (Figure S1), classified by biological process, cellular components and molecular function, respectively.
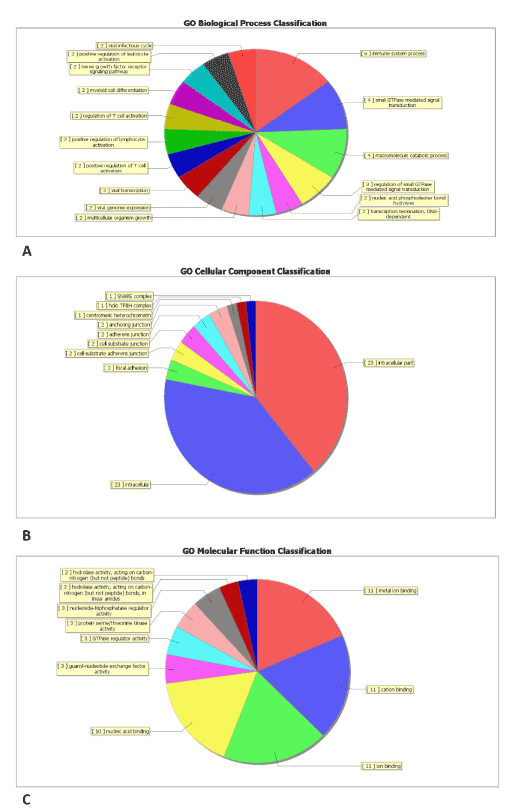
Figure 2. Gene ontology (GO) enrichment analysis of lncRNA-targets. (A) GO analysis of lncRNA-target genes according to biological process, (B) GO analysis of lncRNA-taget genes according to cell component, (C) GO analysis of lncRNA-target genes according to molecular function.
Pathway analysis
Subsequently, we performed the enrichment score analysis (Figure 3) and pathway analysis. We found that 9 pathways through which low shear stress acted on the regulation of cellular function (Table S5). Of these 9 pathways, 2 virus-related (hsa 05162 for measeles, hsa 05161 for hepatitis) and 1 non-small lung cancer-related (has 05223) were excluded from the analysis. Vascular smooth muscle contraction- , adrenergic signaling in cardiomyocytes- and neurotrophin-related signal pathways were also not analyzed from this analysis. Finally, NF-kB, MAPK and Jak-STAT pathways were considered to be as major pathways by which low shear stress regulated endothelial cells, with 9 genes (BDNF, DUSP4, GADD45B, MAP2K5, PLA2G4C, DDX58, GADD45B, TLR4, IL6ST, SPRY1, STAT5A) involved.
According to pathway analysis, two up-stream genes (TLR4 and BDNF) were upregulated (Figure 4 and Table S5). TRL4, one cell membrane located protein, was supposed to be the LSS receptor. As shown in Figure 4, low shear stress significantly upregulated TLR4 and NF-kappa B associated genes. Similarly, low shear stress activated Neurotrophin signaling pathway which also exists in ECs and MAPK pathway both via activation of BDNF. BDNF, a factor that has been reported to be produced by ECs, is a crucial role in the progression of AS.
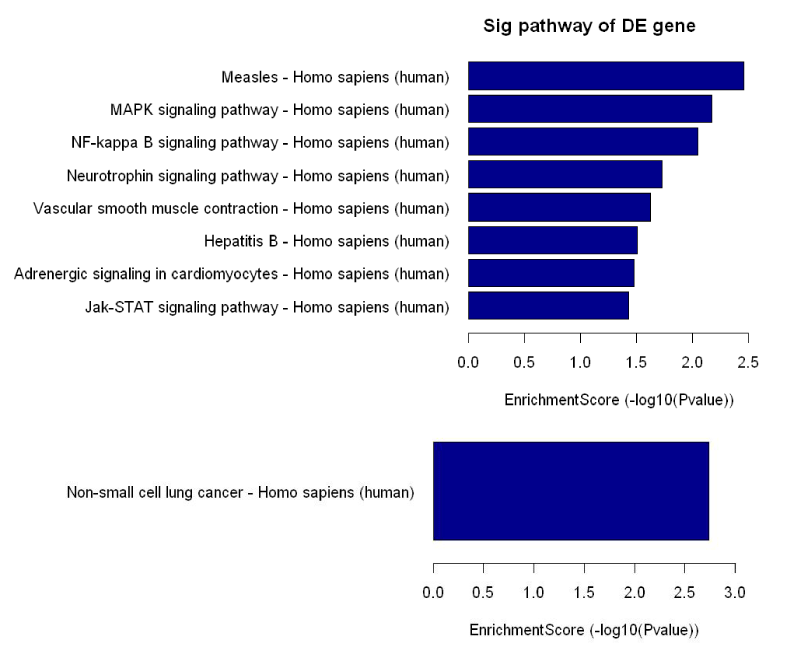
Figure 3. Enrichment score assessment. 8 pathways upregulated by low shear stress (upper row) and 1 pathway downregulated (bottom rom) were assessed according to their scores. Scores indicated the intensity of enrichment.
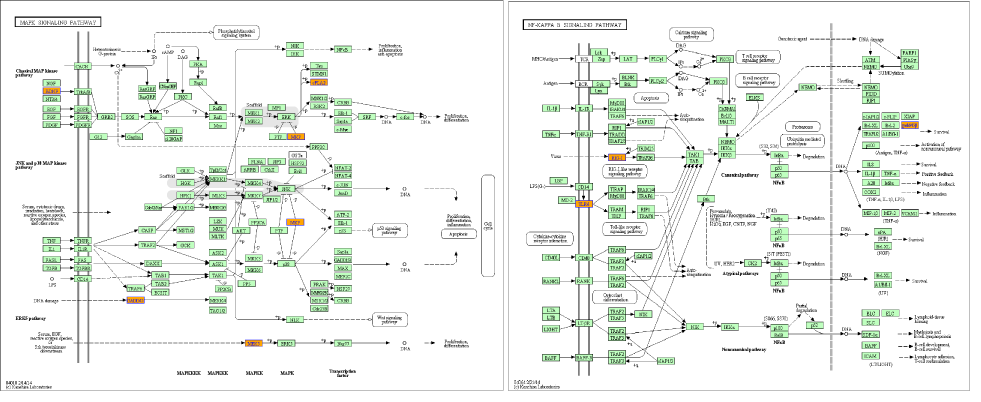
Figure 4. Pathways analysis. According to pathways analysis, two genes (BDNF and TLR4) were upregulated by low shear stress. From KEGG database, BDNF- (left) and TLR4-(right) related pathways were showed.
Our study provides the expression profiles of lncRNA and mRNA in sheared cells (after 120 min) and analyzes the possible mechanisms of cellular responses to low shear stress (2dyn/cm2). The results show that TLR4 and BDNF pathways plays crucial role in low shear stress-induced cellular injury, among 84 paired differentially expressed lncRNA-mRNA.
There is a great interest in the biological function of RNA. According to high-throughput analysis [10,11], regulatory non-coding RNA (ncRNA) can be classified into short (<30 nt in length ), medium (20-300 nt in length) and lncRNA (>200 nt in length). To date, the majority of lncRNA are likely to be functional, but only a relative small proportion has been demonstrated to be biologically relevant, such as regulation of basal transcription mechinery functions in cells [12]. Endothelial cells of different origin express relative high levels of the conserved long noncoding RNAs [13]. Several lncRNAs have been reported to be correlated with atherosclerosis and endothelial cellular dysfunction [14-16]. However, there is a lack of differentially expressed lncRNAs in endothelial cells exposed to low shear stress. Here, we for the first reported that three lncRNAs (C17orf76-AS1, BC079832, and LEF1-AS1) were upregulated and two lncRNAs (XLOC-007914 and RP11-473M20.16) were downregulated after endothelial cells were treated by low shear stress (2.0 dyn/cm2) for 120 min.
C17orf76-AS1, also known as LRRC75A-AS1, dynamically conveyed information from the activated epidermal growth factor and extracellular signal-regulated kinase (EGF/ERK) signaling cascade to directly associated proteins and more distant proteins in the network [17,18], which is consistent with our previous finding that low shear stress activated ERK/MAPK signal pathway [9]. LRRC75A-AS1 and BC079832 mainly regulated zinc finger and BTB domain-containing protein 37(ZBTB37) gene which was involved in the regulation of MAPK signals [17-19].
LEF1-AS1 is another antisense transcript. It regulated the expression of KIAA1731 gene [20,21].
Subsequently, we measured the expression of mRNA in endothelial cells after treated by low shear stress. We found that TLR4 and BDNF, two of major membrane genes, were significantly regulated. TLR4 associates with the activation of NF-κB signaling and plays as monitor of NF-κB [22,23]. Numerous studies have demonstrated that TLR4/NF-κB system participated in the formation and acceleration of AS [24-27]. Yang et al. identified increased TLR4 expression, activated NF-kB signaling in LSS-treated cell and supressed inflammation in TLR4 mutant [24]. Michelsen also reported that genetic deficiency of TLR4 led to a significant reduction in aortic plaque areas in apo-E deficient mice [27]. Triggering TLR4 pathway leads to the activation of NF-κB signaling and subsequent regulation of inflammatory genes. In this study, we found that LSS significantly promoted the expression of TLR4, DDX58, GADD45B, and these genes were involved in NF-κB signaling. Moreover, our study indicated firstly that TLR4 is the membrane located LSS receptor and initiation of LSS-induced endothelium injury.
We assayed BDNF gene in sheared cells and found an increase and we proposed BDNF contributed to the progression of AS. One decade ago, Nakahashi assayed ECs-produced BDNF under high shear stress (24 dynecs/cm2) and found an decrease [28]. Haplo-deficient BDNF-receptor was associated with reduced AS, smooth muscle cells proliferation and collage accumulation in the lesion [29]. These evidences indicate that BDNF is involved in the development of AS. However, ECs derived BDNF depends on endothelial function rather than the artificial stimulations [29]. Others also reported reduced BDNF in AS patients’ serum [30]. Even though the precise role of BDNF is not very clear, it is not advise to deny the association of BDNF with AS, which needs to be further explored.
The present study reported the differential expression of lncRNAs and mRNAs in endothelial cells exposure to low shear stress for 120 minutes. Pathway analysis demonstrated that BDNF and TLR 4 genes played important role in modulating cellular injury induced by low shear stress.
2021 Copyright OAT. All rights reserv
The authors declare that they have no competing interests
CSL designed this study and wrote this manuscript. WZM carried out the shear stress experiments, molecular genetic studies, participated in the sequence alignment and drafted the manuscript. HZY carried out the genetic studies. LB participated in the design of the study and performed the statistical analysis. All authors read and approved the final manuscript.
- Heo KS, Fujiwara K, Abe J (2011) Disturbed-flow-mediated vascular reactive oxygen species induce endothelial dysfunction. Circ J 75: 2722-2730. [Crossref]
- Barton M, Baretella O, Meyer MR (2012) Obesity and risk of vascular disease: importance of endothelium-dependent vasoconstriction. Br J Pharmacol 165: 591-602. [Crossref]
- Reinhart-King CA, Fujiwara K, Berk BC (2008) Physiologic stress-mediated signaling in the endothelium. Methods Enzymol 443: 25-44. [Crossref]
- Koskinas KC, Chatzizisis YS, Antoniadis AP, Giannoglou GD (2012) Role of endothelial shear stress in stent restenosis and thrombosis: pathophysiologic mechanisms and implications for clinical translation. J Am Coll Cardiol 59: 1337-1349. [Crossref]
- Wragg JW, Durant S, McGettrick HM, Sample KM, Egginton S, et al. (2014) Shear stress regulated gene expression and angiogenesis in vascular endothelium. Microcirculation 21: 290-300. [Crossref]
- Dolan JM, Meng H, Sim FJ, Kolega J (2013) Differential gene expression by endothelial cells under positive and negative streamwise gradients of high wall shear stress. Am J Physiol Cell Physiol 305: C854-866. [Crossref]
- Ishibazawa A, Nagaoka T, Yokota H, Ono S, Yoshida A (2013) Low shear stress up-regulation of proinflammatory gene expression in human retinal microvascular endothelial cells. Exp Eye Res 116: 308-311. [Crossref]
- Zhang J, Wang Z, Zhang J, Zuo G1, Li B, et al. (2014) Rapamycin attenuates endothelial apoptosis induced by low shear stress via mTOR and sestrin1 related redox regulation. Mediators Inflamm 2014: 769608. [Crossref]
- Hung T1, Chang HY (2010) Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7: 582-585. [Crossref]
- Shi X, Sun M, Liu H, Yao Y, Song Y (2013) Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 339: 159-166. [Crossref]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS (2009) Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic 8: 407-423. [Crossref]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114: 1389-1397. [Crossref]
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, et al. (2013) Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet 9: e1003588. [Crossref]
- Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, et al. (2013) The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet 22: 4516-4527. [Crossref]
- Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, et al. (2007) Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem 282: 15652-15666. [Crossref]
- Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, et al. (2011) A directed protein interaction network for investigating intracellular signal transduction. Sci Signal 4: rs8. [Crossref]
- Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, et al. (2009) Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J 28: 1271-1282. [Crossref]
- Gibbs R.A, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, et al. (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Rat Genome Sequencing Project Consortium. Nature 428: 493-521
- Kim RN, Kim DW, Choi SH, Chae SH, Nam SH, et al. (2011) Major chimpanzee-specific structural changes in sperm development-associated genes. Funct Integr Genomics 11: 507-517. [Crossref]
- Knorz VJ, Spalluto C, Lessard M, Purvis TL, Adigun FF, et al. (2010) Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif-containing proteins C10orf90 and KIAA1731. Mol Biol Cell 21: 3617-3629. [Crossref]
- Canton G, Chiu B, Chen H, Chen Y, Hatsukami TS, et al. (2013) A framework for the co-registration of hemodynamic forces and atherosclerotic plaque components. Physiol Meas 34: 977-990. [Crossref]
- Cheng C, Tempel D, van Haperen R, van der Baan A, Grosveld F, et al. (2006) Atherosclerotic lesion size and vulnerability are determined by patterns of fluid shear stress. Circulation 113: 2744-2753. [Crossref]
- Wang W, Ha CH, Jhun BS, Wong C, Jain MK, et al. (2010) Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood 115: 2971-2979. [Crossref]
- Yang S, Li R, Tang L, Qu X, Ge G, et al. (2013) TLR4-mediated anti-atherosclerosis mechanisms of angiotensin-converting enzyme inhibitor--fosinopril. Cell Immunol 285: 38-41. [Crossref]
- Fang D, Yang S, Quan W, Jia H, Quan Z, et al. (2014) Atorvastatin suppresses Toll-like receptor 4 expression and NF-κB activation in rabbit atherosclerotic plaques. Eur Rev Med Pharmacol Sci 18: 242-246. [Crossref]
- Yang QW, Mou L, Lv FL, Wang JZ, Wang L, et al. (2005) Role of Toll-like receptor 4/NF-kappaB pathway in monocyte-endothelial adhesion induced by low shear stress and ox-LDL. Biorheology 42: 225-236. [Crossref]
- Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, et al. (2004) Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci U S A 101: 10679-10684. [Crossref]
- Nakahashi T, Fujimura H, Altar CA, Li J, Kambayashi J, et al. (2000) Vascular endothelial cells synthesize and secrete brain-derived neurotrophic factor. FEBS Lett 470: 113-117. [Crossref]
- Norata GD, Pulakazhi Venu VK, Callegari E, Paloschi V, Catapano AL (2012) Effect of Tie-2 conditional deletion of BDNF on atherosclerosis in the ApoE null mutant mouse. Biochim Biophys Acta 1822: 927-935. [Crossref]
- Manni L, Nikolova V, Vyagova D, Chaldakov GN, Aloe L (2005) Reduced plasma levels of NGF and BDNF in patients with acute coronary syndromes. Int J Cardiol 102: 169-171. [Crossref]






