Introduction
Neurodegenerative disorders are often classified as a “multifactorial syndrome” as they share similarities between many genetic, clinical, psychological as well as environmental factors [1,2]. They are highly debilitating clinical conditions that result in progressive neuronal degeneration. Alzheimer Disease (AD) in particular is characterized by progressive neuronal dysfunction and regular decline in cognition and behavior. The cause of AD is broadly classified into two categories: sporadic and familial. The most common form of familial mutations is due to three major genes namely APP, PSEN1 and PSEN2 [3-5]. However, the sporadic form of AD is a complex amalgamation of genetic polymorphisms, environment as well as social lifestyle [6-8]. Although it has been decades since the search for novel biomarkers commenced, there is still no proper diagnosis and treatment for AD [9-11]. Barrett and Hunter’s team report that the lack of efficient treatment for AD could be primarily due to a sort of careless misdiagnosis of the disease by physicians [12,13]. Such errors could be an act of lack of attention in routine medical examinations. The existing health care treatment for AD is symptomatic relief [14,15]. However, it is widely disputed that the altering neurodegenerative patterns actually commence much earlier than the actual clinical manifestation of the disease. Therefore, early detection would not only improve the diagnostic accuracy in the clinics but also aid clinicians to offer better and earlier treatment for cognitive and behavioral problems [16,17] as well as better quality of life and economic outcomes.
State-of-the-art brain imaging technologies provide high-resolution information of structural and functional alterations. Therefore, they offer unprecedented early diagnosis; they also provide the opportunity for regular monitoring of a progressive clinical condition such as AD. Furthermore, imaging techniques aid in tracing the transition between diagnostic states such as Mild Cognitive Impairment (MCI) and AD.
Depending on brain complexity, imaging techniques reveal different dimensions of brain structure and function. They can be broadly classified into three groups namely [18,19]:
- Structural Neuroimaging
- Functional Neuroimaging
- Molecular Neuroimaging
Structural Neuroimaging
Magnetic Resonance Imaging (sMRI), Computed Tomography (CT) and Diffusion Tensor Imaging (DTI) are some of the prominent structural neuroimaging techniques. Structural MRI is widely used to examine the shape, size and structural alterations in the brain regions [20,21]. DTI is an advanced MR technique that helps in understanding structural connectivity between brain regions [22,23]. These techniques primarily help in observable indicators such as “tissue damage” or loss of brain regions as well as measurable indicators such as white or gray matter changes and morphological changes such as cortical thinning [24,25]. These indicators are collectively classified as neuroimaging biomarkers as they are quantitative tracers of the disease progression. Some important neuroimaging biomarkers are listed below:
Atrophy
Brain atrophy is one of the most prominent neuroimaging biomarker for AD. Atrophy refers to the loss of nerves and tissue, which ultimately results in the shrinkage of the brain [26,27]. It has been previously estimated that whole brain atrophy affects 2% of AD patients while the rate of atrophy in normal ageing does not exceed beyond 0.7% per year [28]. According to Frisoni, et al., the earliest MRI based atrophic changes can be detected in entorhinal cortex, hippocampus and cingulate cortex resulting in early memory dysfunction [29,30].
Cortical thinning
Many histopathological studies have proposed that AD are often related to damage of specific cortical layers such as neocortex and entorhinal cortex [31,32]. The latest MRI techniques still are not capable of examining individual layers of the cortex. However, there are many semi-automated surface reconstruction tools such as FreeSurfer, 3D MPRAGE that aid in cortex examination [33-35].
Fractional Anisotropy
DTI techniques tracts the water diffusion in various tissues which provide vital information. They help in measuring the structure of white matter as well as fiber connectivity within and across brain regions [36-38]. Fractional anisotropy is a numerical measure of fiber integrity. This index is sensitive enough to detect the white matter degradation in aging and other neurodegenerative diseases [39-41] (Figure 1).
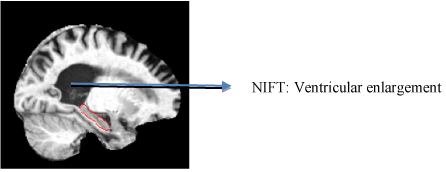
Figure 1. T13D MP-RAGE scan of structural neuroimaging. This figure illustrates the image scan of an AD patient with ventricular enlargement annotated using NIFT terminology
Functional Neuroimaging
Functional neuroimaging techniques help in determining the severity of brain injury which reflects on the cognitive and behavioral changes amongst patients [42,43]. The most commonly used functional neuroimaging techniques include functional MRI, Positron Emission Tomography (PET), Single Photon Emission Computed Tomography (SPECT), Electroencephalography (EEG) and Transcranial Magnetic Simulation (TMS) [44,45] and MR Spectroscopy (MRS).
MRS is a widely used non-invasive imaging technique that helps in measuring the metabolites found in brain tissues. It also facilitates in measuring the chemical composition of tissues such as myo-inositol, choline, n-acetyl aspartate as well as choline. The advanced MRS imaging techniques help in identifying patients much ahead of the clinical onset of AD [46,47].
Brain Glucose metabolism
Recent advancements in functional imaging studies have contributed significantly to identification of patterns amongst patients, who are at the risk of developing AD [48,49]. The earliest PET imaging abased studies were used to detect altering glucose metabolic changes amongst patients who were at a genetic risk of developing AD [50-52]. PET-based radioisotopes such as oxygen (015) aid in tracing changes in cerebral blood flow which are often caused due to increased neuronal activity [53-55]. Similarly, [18 F] fludeoxyglucose-positron emission tomography (FDG-PET) detects bilateral temporoparietal hypometabolism [56-58]. They have been widely used as a diagnostic differential biomarker discriminating between patients with AD dementia and vascular dementia [59,60]. Another radioisotope based biomarker that is widely used in diagnostic studies is C-labeled Pittsburgh Compound-B ([(11)C]PIB). The increased binding potential of PiB was found to be common amongst MCI patients whereas decreased FDG uptake was observed only with patients with AD, thus serving a crucial diagnostic biomarker [61,62] (Figure 2).
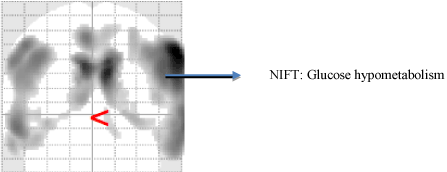
Figure 2. Functional Neuroimaging using FDG [18] PET. This figure represents an AD patient scan with excessive excessive glucose hypometabolism
Perfusion
Imaging techniques such as SPECT and DTI enable early detection of hypoperfusion in the white matter and cortex [63,64]. Abnormal cerebral perfusion are clear indicators of diagnostic transition from MCI to AD [65,66]. Borroni and Chao et al., has demonstrated patterns of hypoperfusion in parietal, temporal and posterior cingulate cortex in all those patients who are progressing from MCI to AD [67,68]. Another study performed by Caroli et al., compared three diagnostic groups namely CN, MCI and AD. The outcome of this study reported that hippocampal hypoperfusion pattern was found across patients with amnestic MCI in transition to AD [69] (Figure 3).
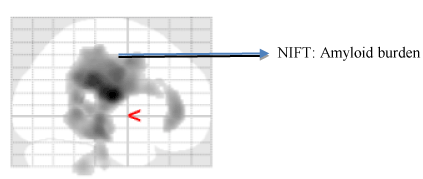
Figure 3. Molecular neuroimaging using [18F] AV 45 PET. This figure displays the amyloid burden in coronal, sagittal regions of AD patient
Emerging combinatorial biomarkers for AD
Clinical neuroimaging biomarkers are useful resources for AD diagnosis. However, the characteristics of these imaging biomarkers are not yet adequate for diagnosis of patients at an individual level. This is largely due to the lack of longitudinal imaging data [70,71]. Combining known genetic biomarkers with imaging data could improve the prediction pattern across all patients [72-74]. Neuroimaging genetics is an emerging field in which quantitative phenotypic features from brain imaging are used as readout to inspect the role of genetic variation in brain function [75,76].
Large scale GWAS studies have contributed to the identification of many risk mutations associated with AD such as CLU, PICALM, BIN1, CR1 and so on [77-79]. These studies have created a substantial shift in the mundane AD detection through standard cognitive tests. Of all the above mentioned genes, CLU is the most significant gene used in combinatorial imaging analysis. The risk variant rs11136000 have been associated with reduction in hippocampal volume in patients with Late Onset Alzheimer Disease (LOAD) [80-82]. Apart from CLU, the risk variant rs541458 of PICALM was found to be associated with CSF Abeta 42 levels [83-85]. Similarly, large scale initiatives across the globe have already started investing in the direction of combining genetic and imaging derived biomarkers for better AD diagnosis (Table 1).
Table 1. Represents a sample of cohort-based studies done using combinations of biomarkers for early AD detection
Study type |
Cohort |
Snps |
Imaging readouts |
Outcome |
Voineskos, et al. [86] |
Philadelphia Neurodevelopmental cohort |
rs12148337 |
White matter fractional anisotropy |
The mutation had a polygenic risk score with white matter FA in schizophrenic population |
Louwersheimer, et al. [87] |
Amsterdam Dementia Cohort |
rs2070045-G (SORL1) |
Hippocampal atrophy |
SORL1 SNP rs2070045-G allele was related to CSF-tau and hippocampal atrophy, 2 endophenotype markers of AD, suggesting that SORL1 may be implicated in the downstream pathology in AD. |
Benussi, et al. [88] |
Brescia Cohort |
Leu271LeufsX10 (PGRN) |
Multiple System Atrophy |
PGRN mutations were shown in familial FTLD, 75% in Corticobasal syndrome |
Morris, et al. [89] |
Cohort of 355 stroke survivors |
rs1799983, p.Asp298Glu |
Cerebral perfusion |
The presence of TT genotype increased risk of incident dementia compared with GG genotype; hazard ratio, 3.14 (95% confidence interval, 1.64-5.99; p = 0.001). |
Schuur M, et al. [90] |
Dutch family based cohort |
rs1699102, rs3824968, rs2282649, rs1010159 |
Microbleeds |
The association of SORL1 with microbleeds suggests that the amyloid cascade is involved in the aetiology of microbleeds in populations with hypertension. |
Inkster, et al. [91] |
AD cohort |
rs10868366 |
Gray matter volume |
The greater effect size in AD patients also suggests that the GG genotype could be a risk factor for the expression of cognitive deficits in AD. |
Lyall, et al. [92] |
Lothian Birth Cohort |
rs10524523 |
Hippocampal volume |
|
Assareh, et al. [93] |
Longitudinal Sydney Older Persons Study |
rs4935774-T, rs2298813-G, rs1133174-G |
Hippocampal atrophy |
The most common haplotype (H1), comprising rs4935774-T, rs2298813-G, and rs1133174-G alleles (T/G/G) was associated with whole brain atrophy in both males and females (p=0.012 & p=0.013; respectively). |
Oliveira-Filho, et al. [94] |
Boston Cohort |
rs20417 |
White matter hyperintensity volume |
rs20417 polymorphism was associated with increased WMHv (P = .037),not cardioembolic stroke patients. |
Large scale initiatives on neuroimaging and genetics
Here, we summarize the various initiatives that are focusing on integrating multi-scale data such as imaging and genetics for efficient diagnosis and treatment.
ADNI
ADNI is considered as one of the biggest ongoing multicenter study for developing longitudinal clinical, imaging, genetic and neuropsychological biomarkers for early detection of AD. The initial phase (ADNI-1) study had the greatest enrollment of participants comprising of 400 early MCI subjects, 200 AD and 200 Controls. Owing to its success, the study was further extended into the next phase (ADNI-2) with additional 550 participants. This study aimed at developing a standardized protocol for data integration and collection for MRI, PET and CSF biomarkers in a global environment [95,96]. The outcome of this study produced interesting hypotheses which went beyond conventional understanding of the AD pathology. One of the earlier studies demonstrated that image derived biomarkers such as “atrophy” and “hypometabolism” exhibited a pattern based on the disease progression and severity [97,98]. Many successive studies also demonstrated the importance of CSF biomarkers, PET based biomarkers as early indicators of pre-clinical AD [99-101]. Another sister initiative of ADNI is called ADNI Genetics Core, which provides the possibility for researchers to estimate the genetic alterations using imaging features for understanding disease progression over time [102-104].
The European Alzheimer's disease Neuroimaging Initiative (E-ADNI)
The overall goal of the E-ADNI initiative was to apply the standardized protocol of collecting images, genetics, and clinical as well as psychological data by adapting the European Centers of the Alzheimer ’s disease Consortium (EADC). This initiative was propelled to encourage the academic EADC centers to adopt the ADNI protocol for enrolling participants [105,106].
The Italian Alzheimer’s Disease Neuroimaging Initiative (I-ADNI)
The I-ADNI initiative was launched in succession to US-ADNI study for validating the acquisition and processing protocol of structural MRI scans obtained from different clinics across Italy by following the procedure from the original ADNI study [107,108].
The Australian Imaging Biomarkers and Lifestyle Study of Aging (AIBL)
The AIBL (https://aibl.csiro.au/about/) initiative consists of 1,200 Australian participants who were longitudinally assessed for over 5 years. This study was launched in 2006 to identify biomarkers, cognitive assessments, genotype, biomarkers such as APOE, social and health factors for monitoring AD progression and early AD treatment. The AIBL initiative has given rise to lot of insights such as AD patients are prone to be more anemic than patients with MCI [109,110]. Participants enrolled in this initiative are continuously assessed every 18 months for any clinical indication of the disease.
EPAD
EPAD (http://ep-ad.org/) stands for European Prevention of Alzheimer’s Dementia Consortium. It is a major European initiative for developing systematic and flexible approaches to clinical trials of drugs for preventing Alzheimer’s dementia. The adaptive trial design in EPAD promises a faster and low cost drug production in the market. The imaging protocol of EPAD is adapted from the AMYPAD initiative which brings together the academic and private research groups for PET based studies to explore amyloid-beta as a therapeutic marker for AD [111,112].
AMYPAD
AMYPAD (http://www.amypad.eu/) stands for Amyloid Imaging to Prevent AD. This project was initiated to investigate the beta-amyloid biology through PET scans from pre-symptomatic population as a diagnostic and therapeutic biomarker for AD. The AMYPAD project is funded by the Innovative Medicine Initiative (IMI) program and will run initially over 5 years. In the course of this project, patients susceptible to AD will be scanned for beta-amyloid through PET imaging. The initiative aims at improving the diagnostic standards for AD treatment and prevention (http://www.alzheimer-europe.org/News/EU-projects/Thursday-17-December-2015-AMYPAD-project-progresses-to-second-stage-of-applications-for-IMI2-Call-5).
PPMI
The Parkinson’s Progressive Markers Initiative (PPMI) (http://www.ppmi-info.org/) is an observational longitudinal clinical study designed for examining patients with Parkinson’s Disease (PD), healthy controls and also participants who have higher chances of developing PD. This study comprises of 1000 participants examined regularly over 8 years and the patients are enrolled in 33 clinical sites in the US, in Europe, and in Asia. The PPMI data resource comprises of clinical and behavioral assessments, imaging data and biospecimen such as CSF, DNA, RNA, plasma, urine and cell line samples. PPMI is funded by the Michael J Fox Foundation in collaboration with 18 biotech and pharmaceutical companies [113].
ENIGMA
ENIGMA (http://enigma.ini.usc.edu/) stands for Enhancing NeuroImaging Genetics Through Meta-Analysis. This consortium is an effort towards bringing researchers from diverse domains such as imaging genomics, neurology and psychiatry together to understand brain structure and function through MRI, DTI, fMRI, genetic as well as patient data. This study has so far analyzed 12,826 subjects. The preliminary project of ENIGMA was to identify common genetic variants in hippocampal or intracranial volume using Genome Wide Association Studies (GWAS). ENIGMA2 was the next project to explore genetic variants associated with subcortical volumes and ENIGMA-DTI was designed to explore genetic variants associated with white matter microstructures. Apart from meta-analysis based studies, the consortia are also focusing on understanding, how psychiatric conditions such as schizophrenia, bipolar disorder, depression affect brain functionality [114,115].
NeuroImage
NeuroImage (http://www.neuroimage.nl/) is an International Multiscale Attention-Deficit/Hyperactivity Disorder (ADHD) Genetics Initiative (IMAGE) funded by the National Institute of Mental Health. The goal of the study is to gather and analyze endophenotypic, phenotypic and genetic information about ADHD. This study is based on a collection of 5,578 subjects from 8 European countries. In the course of this project, structural and functional MRI scans are performed on patients, along with neuropsychological assessments and GWAS analysis in order to detect functional abnormalities underlying ADHD [116,117].
Initiatives such as ADNI and PPMI have largely invested in systematically harvesting genetic and imaging data. Studies like ADNI and PPMI form the basis for the association of imaging readouts with genetic variation information and may facilitate the generation of hypotheses about mechanistic links between genes and imaging features.
Mining links between neuroimaging readouts and molecular processes from literature
High-throughput imaging technologies have been employed to understand the molecular mechanisms underlying clinical conditions. Such efforts have led to the identification of novel biomarkers for all disease domains, especially AD [118,119]. However, the rapid growth of the literature around these combinatorial studies has made it increasingly difficult to aggregate and mine the reported findings [120]. Obviously, new technologies enabling automated text processing (“text-mining”) may help to retrieve relevant documents and to extract relevant knowledge from text.
Ontologies and terminologies
One of the most efficient ways to address the challenge of unstructured information mining is with the efficient usage of ontologies and controlled terminologies. Ontologies are formal representations of knowledge that can represent entire research domains. They are helpful when concepts need to be shared across research communities in an unambiguous fashion. This is very crucial as it enables different research groups to communicate with each other without misinterpretation of the biological context [121-123]. Ontologies do also facilitate the exchange of data and knowledge between machines; they are in fact readable by both, human experts and machines. When transformed into terminologies (dictionaries), they can readily be integrated into text-mining systems and are very useful for information extraction and knowledge representation. Furthermore, ontologies bear the potential to enable automated reasoning over knowledge representations [124,125].
Existing ontologies in the field of neuroimaging
Similar to other biological domains, the field of neuroimaging research has advanced semantically by generating various terminologies and ontologies in the past. Some of the more widely recognized neuroimaging ontologies are listed below:
Quantitative Imaging Biomarker Ontology (QIBO)
QIBO ontology was developed to standardize quantitative imaging biomarkers for better therapeutic intervention. This ontology consists of 488 terms and they consist of classes such as imaging agent, imaging instrument or biological intervention. QIBO represents concepts across several fields, including imaging physics and biology [126].
Magnetic Resonance Imaging Ontology (MRIO)
This ontology captures all concepts needed to describe the outcome of MRI scans. It has been designed to overcome the heterogeneity in MRI readouts. The authors mainly capture measured data coming from T1, T2, tissue as well as other factors, such as temperature. The MRIO ontology focusses mainly on two MRI representations namely MRI simulators and DICOM images and conceptualize all possible terms that can be observed using these scanned images [127,128].
NeuroLog
The NeuroLog consortium was established in the year 2006 for sharing and reusing data and tools for neuroimaging studies. This software architecture aids in efficient integration of neuroimaging data and tools from various neuroimaging research centers. This consortium also takes charge of the autonomous data management from each center to maintain the confidentiality of the neuroimaging data. Furthermore, the usage of semantically annotated tools inbuilt in the system architecture provides better standardization of neuroimaging datasets and therefore offers better accessibility through the federated schema based ontology [129,130].
NeuroImage Feature Terminology (NIFT)
Although there are so many ontologies established in the area of neuro-imaging, there is still a lack of a terminology which facilitates a systematic representation and retrieval of measured indices with high relevance for neurodegenerative diseases. All the existing ontologies represent what the imaging scan capture, but they do not contain concepts that link imaging readouts to disease pathology. Motivated by the apparent need for such a terminology, we have developed NIFT, the “neuro-image feature terminology”. NIFT represents a wide spectrum of terms linked to radiological, neuropsychological as well as measured indices highly relevant to neurodegenerative diseases (e.g. AD and PD) [131]. The NIFT terminology comprises highly generic concepts describing common neuroimaging features, but at the same time it is very specific and represents disease-centric pathological measures used in imaging scans in the domain of Alzheimer ’s disease and Parkinsonism. NIFT can act as a potential resource to capture molecular as well as clinical readouts, which are crucial in bridging these two domains as well as retrieving relevant documents which can be further used in a multi-layered disease models. As such, NIFT is well suited to support the identification of novel mechanisms underlying the etiology of AD and PD.
Retrieval of relevant publications using the nift terminology
The main purpose for developing ontologies and terminologies is to retrieve relevant publications and automatically extract relevant information from the literature. To enable specific retrieval and information extraction in the imaging domain, we integrated the NIFT terminology into our in-house text-mining system SCAIView [132,133]. SCAIView was developed at Fraunhofer SCAI to enable biologists and clinical researchers to perform semantic search and information extraction from the scientific literature. A free version of this literature mining environment, SCAIView academia, allows free access to the semantically annotated PubMed abstracts. For PubMed Central (PMC) full text publications, SCAIView allows a full-text search as well. We have integrated NIFT in SCAIView and used the system to systematically retrieve relevant documents containing useful information on imaging readouts linked to molecular entities. The resulting literature corpus was then used for mechanistic modeling purposes.
Mechanistic modeling of neuroimaging indices
We wanted to understand the significance of a measured index obtained from imaging techniques and their association with clinical tests to improve the prediction an underlying neurodegenerative disease, in this case, AD. For this, we performed an optimized search query using our literature-mining environment SCAIView.
We used the query “[Neuroimaging Feature]) AND [MeSH Disease: “Alzheimer Disease“]) AND [Alzheimer Ontology Node: “Evaluation“]) AND [BRCO]) AND [PTS]) AND [Organism: ”Homo sapiens“]” to retrieve relevant publications that comprises disease-specific terms, brain region and cell-type information (BRCO) and that comprise pathway mentions (PTS). The Alzheimer Ontology (ADO) concept “evaluation” provides a wide spectrum of entities that describe various clinical tests that are significant for diagnosing AD. Once the articles were retrieved, we tried to model them in order to identify underlying the molecular mechanisms.
Mechanistic modeling of neuroimaging features with molecular pointers
One major motivation to develop the NIFT terminology was to support the generation of cause-and-effect models in the area of neurodegenerative diseases. With the integration of imaging features in cause-and-effect models, we hope to bridge between the molecular level (genome, pathways) and the macroscopic anatomical level of brain structures such as brain regions and the entire organ.
Using the query described above, we generated a literature corpus highly enriched for mentions of interesting imaging features together with interesting molecular processes. One of the resulting models that link imaging features to the molecular pathophysiology of AD deals with the influence of cerebral blood flow on cognitive impairment in AD. The overall workflow applied is shown in Figure 4.
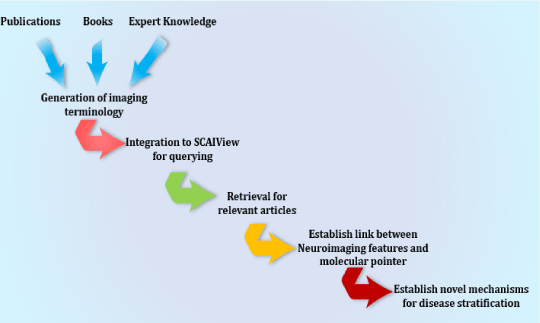
Figure 4. Schematic representation of the workflow to extract links between imaging features and molecular mechanisms in a disease context
NIFT Application Example
Hypothetical model for linking high-level cerebral blood flow with molecular processes:
The scientific community has long been interested in the vascular biology, in which the human physiology is represented as large and small blood vessels which might play a role in AD progression [134,135]. Although clinical studies conducted on AD patients reveal substantial evidences of vascular lesion being the biggest factor of AD, the fundamental understanding of the molecular mechanism behind that remains unexplained [136,137]. Therefore, here we establish our first hypothetical model that links high level complex biology such as cerebral blood flow with molecular processes. This model is highly putative due to the lack of experimental validation and lack of clinical resources to support the hypothesis.
AD is highly diverse and complex in terms of the various cellular and molecular players that together result in the disease pathology. Apart from the molecular deposits such as plaques and tangles, increasing supporting evidences on the role of vascular abnormalities in AD pathology, so much so that these co-morbid conditions are classified under the term “vascular dementia” [138-140].
The links between vascular lesions and cognition impairment are based on observations that have been captured using advanced neuroimaging techniques such as SPECT [141,142]. By using radioisotopic tracers, depletion of blood flow can be traced by reduced glucose consumption in a particular brain region [143,144]. Apart from SPECT, MRI tensors are constantly tuned to detect early neoplasms and altered blood flow detection with high-resolution quality [145,146].
Hypothetical mechanism for Cerebral Blood Flow in AD
SIRT1 stands for “Silient Information Regulator 2 homolog 1”. In general, their role is to maintain cellular functions and promote longevity of the cells in humans as well as other model organisms [147,148]. Sirtuins have been reported to protect the brain from infarction by regulating the blood flow to all parts of the brain, especially the cerebral region [149-151]. In normal conditions, SIRT1 has been reported to play a protective role by enhancing the non-amyloidogenic cleavage of amyloid-beta protein (APP) through NF-kb inhibition. The inhibition of NF-kb contributes to the clearance of amyloid plaques from the brain [152,153]. However, in case of AD, SIRT1 genes are reported to be under expressed which in turn activates the accumulation of amyloid beta in cerebral cortex through NF-Kb activation. The accumulation of APP in the cerebral region could further lead to the depletion of nutrients such as oxygen from the blood, resulting in the inhibition of cerebral blood flow. Lack of oxygen and other nutrients to the brain, various mental and psychiatric abnormalities and could lead to cognitive impairment [154,155].
Also, we hypothesize that the overexpression of SIRT1 co-activates a regulator, which transcribes ADAM10 [156-158]. This could trigger ADAM10 to partially compete with the gamma-secretase for APP fragment resulting in the activation of Notch signaling pathway which is well-known for neuronal repair [159-161]. However, in case of AD ADAM mutant Q170H and R181G does not compete with alpha-secretase, therefore the beta-secretases accumulate in the brain resulting in impaired cerebral blood flow [162-164].
Another plausible mechanism of reduced cerebral blood flow is due to APOE activity. Increased expression of APOE also facilitates the molecular interaction between amyloid beta and Butyrylcholineesterase (BCHE) gene which results in the formation of a complex BCHE-Abeta-APOE (BaβA) complex [165-167]. This complex alters the structure of BCHE which accelerates the catalytic activity of the enzyme. This results in the formation of amyloid plaques [168-170] as seen Figure 5. Increased expression of APOE also disrupts the neuronal activity in the hippocampus resulting in atrophy. Hippocampal atrophy is also one of the causative factor of cognitive decline in AD [171-173].
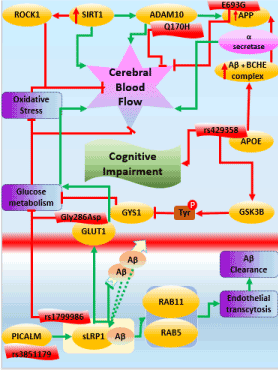
Figure 5. Mechanistic modeling of cerebral blood flow in AD
Apart from the well-known genes of AD, recently, PICALM gene has been emerging as a potential AD candidate. PICALM plays a crucial role in intracellular trafficking of endothelial proteins resulting in endocytosis. The protective allele of PICALM, rs3851179 facilitates the amyloid beta clearance through endocytosis [174-176]. LRP1 is another crucial protein whose major function is cholesterol transport and transcytosis of various molecules including amyloid beta across the BBB [177-179]. As PICALM plays a major role in the internalization of the endothelial proteins, it also internalizes the sLRP1 and amyloid-beta complex by trafficking through two other proteins Rab5 and Rab11. These further results in amyloid transcytosis and clearance from entering the BBB [180-182]. Also, LRP1 activates another protein called GLUT1 which is another major glucose transporter across the BBB [183-185]. During normal conditions, there is a free flow of glucose and other nutrients across BBB. However, during AD, GLUT function is altered by Gly286Asp resulting in inhibition of glucose metabolism [186-188].
Here, we have demonstrated a hypothetical mechanism around cerebral blood flow in AD. We call this model as “putative” and “hypothetical” because they lack individual causal proof and substantial experimental validation. The overall workflow of the altered regulation of cerebral blood flow can be seen in Figure 5.
References
- Iqbal K, Grundke-Iqbal I (2010) Alzheimer's disease, a multifactorial disorder seeking multitherapies. Alzheimers Dement 6: 420-424. [crossref]
- Carmo Carreiras M, Mendes E, Jesus Perry M, Paula Francisco A, Marco-Contelles J (2013) The multifactorial nature of Alzheimer’s disease for developing potential therapeutics. Current topics in medicinal chemistry 13: 1745-1770. [crossref]
- Zekanowski C, Styczynska M, Peplonska,B, Gabryelewicz T, Religa D, et al. (2003) Mutations in presenilin 1, presenilin 2 and amyloid precursor protein genes in patients with early-onset Alzheimer's disease in Poland. Experimental neurology 184: 991-996. [crossref]
- Ryan NS, Rossor MN (2010) Correlating familial Alzheimer's disease gene mutations with clinical phenotype. Biomark Med 4: 99-112. [crossref]
- Weggen S, Beher D (2012) Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer's disease. Alzheimer's research & therapy 4: 1. [crossref]
- Combarros O, Alvarez-Arcaya A, Sanchez-Guerra M, Infante J, Berciano J (2002) Candidate gene association studies in sporadic Alzheimer’s disease. Dementia and geriatric cognitive disorders 14: 41-54. [crossref]
- Bekris LM, Yu CE, Bird TD, Tsuang DW (2010). Review article: genetics of Alzheimer disease. Journal of geriatric psychiatry and neurology 23: 213-227. [crossref]
- Makovac E, Cercignani M, Serra L, Torso M, Spanò B, et al. (2016) Brain Connectivity Changes in Autosomal Recessive Parkinson Disease: A Model for the Sporadic Form. PloS one 11: e0163980. [crossref]
- Antila K, Lötjönen,J, Thurfjell L, Laine J, Massimini M, et al. (2013) The PredictAD project: development of novel biomarkers and analysis software for early diagnosis of the Alzheimer's disease. Interface focus 3: 20120072. [crossref]
- Galvin JE (2012) Optimizing diagnosis and management in mild-to-moderate Alzheimer’s disease. Neurodegener Dis Manag 2: 291-304. [crossref]
- Thaipisuttikul P, Galvin JE (2012) Use of medical foods and nutritional approaches in the treatment of Alzheimer's disease. Clin Pract (Lond) 9: 199-209. [crossref]
- Barrett JJ, Haley WE, Harrell LE, Powers RE (1997) Knowledge about Alzheimer disease among primary care physicians, psychologists, nurses, and social workers. Alzheimer Disease & Associated Disorders 11: 99-106. [crossref]
- Hunter CA, Kirson NY, Desai U, Cummings AKG, Faries DE, et al. (2015) Medical costs of Alzheimer's disease misdiagnosis among US Medicare beneficiaries. Alzheimer's & Dementia 11: 887-895. [crossref]
- Yiannopoulou KG, Papageorgiou SG (2013) Current and future treatments for Alzheimer's disease. Ther Adv Neurol Disord 6: 19-33. [crossref]
- Galimberti D, Scarpini E (2011) Disease-modifying treatments for Alzheimer's disease. Ther Adv Neurol Disord 4: 203-216. [crossref]
- Nestor PJ, Scheltens P, Hodges JR (2004) Advances in the early detection of Alzheimer's disease. Nat Med 10 Suppl: S34-41. [crossref]
- Small GW (2000) Early diagnosis of Alzheimer's disease: update on combining genetic and brain-imaging measures. Dialogues Clin Neurosci 2: 241-246. [crossref]
- Rudin M (2009) Noninvasive structural, functional, and molecular imaging in drug development. Current opinion in chemical biology 13: 360-371. [crossref]
- Saykin A, Shen L, Risacher S, Swaminathan S, Kim S, et al. (2012) Structural, functional and molecular imaging as intermediate phenotypes for studies of candidate genes, pathways and GWAS in Alzheimer's disease. Alzheimer's & Dementia: The Journal of the Alzheimer's Association 8: 232. [crossref]
- Fallon N, Alghamdi J, Chiu Y, Sluming V, Nurmikko T, et al. (2013) Structural alterations in brainstem of fibromyalgia syndrome patients correlate with sensitivity to mechanical pressure. NeuroImage 3: 163-170. [crossref]
- Alexander-Bloch A, Giedd JN, Bullmore E (2013) Imaging structural co-variance between human brain regions. Nature Reviews Neuroscience 14: 322-336. [crossref]
- Mori S, Zhang J (2006) Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51: 527-539. [crossref]
- Kalinosky BT, Schindler-Ivens S, Schmit BD (2013) White matter structural connectivity is associated with sensorimotor function in stroke survivors. NeuroImage 2: 767-781. [crossref]
- Kharitonova M, Martin RE, Gabrieli JD, Sheridan MA (2013) Cortical gray-matter thinning is associated with age-related improvements on executive function tasks. Developmental cognitive neuroscience 6: 61-71. [crossref]
- Ouyang X, Chen K, Yao L, Hu B, Wu X, et al. (2015) Simultaneous changes in gray matter volume and white matter fractional anisotropy in Alzheimer’s disease revealed by multimodal CCA and joint ICA. Neuroscience 301: 553-562. [crossref]
- Peters R (2006) Ageing and the brain. Postgrad Med J 82: 84-88. [crossref]
- Palop JJ, Mucke L (2016) Network abnormalities and interneuron dysfunction in Alzheimer disease. Nature Reviews Neuroscience 17: 777–792. [crossref]
- Sluimer JD, van der Flier WM, Karas GB, Fox NC, Scheltens P, et al. (2008) Whole-brain atrophy rate and cognitive decline: longitudinal MR study of memory clinic patients 1. Radiology 248: 590-598. [crossref]
- Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, et al. (2008) Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain 131: 3266-3276. [crossref]
- Frisoni GB, Laakso MP, Beltramello A, Geroldi C, Bianchetti A, et al. (1999) Hippocampal and entorhinal cortex atrophy in frontotemporal dementia and Alzheimer's disease. Neurology 52: 91-100. [crossref]
- Kersaitis C, Halliday GM, Kril JJ (2004) Regional and cellular pathology in frontotemporal dementia: relationship to stage of disease in cases with and without Pick bodies. Acta Neuropathol 108: 515-523. [crossref]
- Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT (2011) Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med 1: a006189. [crossref]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, et al. (2007) Different regional patterns of cortical thinning in Alzheimer's disease and frontotemporal dementia. Brain 130: 1159-1166. [crossref]
- Lüsebrink F, Wollrab A, Speck O (2013) Cortical thickness determination of the human brain using high resolution 3T and 7T MRI data. Neuroimage 70: 122-131. [crossref]
- Redolfi A, Manset D, Barkhof F, Wahlund LO, Glatard T, et al. (2015) Head-to-head comparison of two popular cortical thickness extraction algorithms: a cross-sectional and longitudinal study. PloS one 10: e0117692. [crossref]
- Fonteijn HM, Norris DG, Verstraten FA (2008) Exploring the anatomical basis of effective connectivity models with DTI-based fiber tractography. Journal of Biomedical Imaging 2008: 3. [crossref]
- Soldner J, Meindl T, Koch W, Bokde AL, Reiser MF, et al. (2012) [Structural and functional neuronal connectivity in Alzheimer's disease: a combined DTI and fMRI study]. Nervenarzt 83: 878-887. [crossref]
- Acosta-Cabronero J, Nestor PJ (2014) Diffusion tensor imaging in Alzheimer's disease: insights into the limbic-diencephalic network and methodological considerations. Frontiers in aging neuroscience 6: 266. [crossref]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, et al. (2007) Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer disease. Neurology 68: 13-19. [crossref]
- Bennett IJ, Madden DJ, Vaidya CJ, Howard DV, Howard JH (2010) Age-related differences in multiple measures of white matter integrity: A diffusion tensor imaging study of healthy aging. Human brain mapping 31: 378-390. [crossref]
- Welton T, Kent D, Constantinescu CS, Auer DP, Dineen RA (2014) Functionally relevant white matter degradation in multiple sclerosis: a tract-based spatial meta-analysis. Radiology 275: 89-96. [crossref]
- Ng TS, Lin AP, Koerte IK, Pasternak O, Liao H, et al. (2014) Neuroimaging in repetitive brain trauma. Alzheimers Res Ther 6: 10. [crossref]
- Beauregard M (2014) Functional neuroimaging studies of the effects of psychotherapy. Dialogues Clin Neurosci 16: 75-81. [crossref]
- Johnson KA, Fox NC, Sperling RA, Klunk WE (2012) Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med 2: a006213. [crossref]
- Niethammer M, Feigin A, Eidelberg D (2012) Functional neuroimaging in Parkinson's disease. Cold Spring Harb Perspect Med 2: a009274. [crossref]
- Chen JG, Charles HC, Barboriak D, Doraiswamy PM (2000) Magnetic resonance spectroscopy in Alzheimer's disease: focus on N‐acetylaspartate. Acta Neurologica Scandinavica 102: 20-26. [crossref]
- Mandal PK (2007) Magnetic resonance spectroscopy (MRS) and its application in Alzheimer's disease. Concepts in Magnetic Resonance Part A 30: 40-64 [crossref]
- Ferreira LK, Busatto GF (2011) Neuroimaging in Alzheimer's disease: current role in clinical practice and potential future applications. Clinics (Sao Paulo) 66 Suppl 1: 19-24. [crossref]
- Mak E, Su L, Williams GB, O'Brien JT (2014) Neuroimaging characteristics of dementia with Lewy bodies. Alzheimers Res Ther 6: 18. [crossref]
- Small GW (1996) Neuroimaging and genetic assessment for early diagnosis of Alzheimer's disease. J Clin Psychiatry 57 Suppl 14: 9-13. [crossref]
- von Leden RE, Selwyn RG, Jaiswal S, Wilson CM, Khayrullina G, et al. (2016) 18 F-FDG-PET imaging of rat spinal cord demonstrates altered glucose uptake acutely after contusion injury. Neuroscience letters 621: 126-132. [crossref]
- Schroeder FA, Chonde DB, Riley MM, Moseley CK, Granda ML, et al. (2013) FDG-PET imaging reveals local brain glucose utilization is altered by class I histone deacetylase inhibitors. Neuroscience letters 550: 119-124. [crossref]
- Owler BK, Momjian S, Czosnyka Z, Czosnyka M, Péna A, et al. (2004) Normal Pressure Hydrocephalus and Cerebral Blood Flow: A PET Study of Baseline Values. Journal of Cerebral Blood Flow & Metabolism 24: 17-23. [crossref]
- Grüner JM, Paamand R, Højgaard L, Law I (2011) Brain perfusion CT compared with 15 OH 2 O-PET in healthy subjects. EJNMMI research 1: 1. [crossref]
2021 Copyright OAT. All rights reserv
- Su Y, Vlassenko A, Blazey T, Ances B, Snyder A, et al. (2014) Comparison of cerebral blood flow measurement obtained from simultaneously acquired ASL and O15-water PET. J Nucl Med 55: 207-207. [crossref]
- Womack KB, Diaz-Arrastia R, Aizenstein HJ, Arnold SE, Barbas NR, et al. (2011) Temporoparietal hypometabolism in frontotemporal lobar degeneration and associated imaging diagnostic errors. Archives of Neurology 68: 329-337. [crossref]
- Newberg AB, Arnold SE, Wintering N, Rovner BW, Alavi A (2012) Initial clinical comparison of 18F-florbetapir and 18F-FDG PET in patients with Alzheimer disease and controls. J Nucl Med 53: 902-907. [crossref]
- Marcus C, Mena E, Subramaniam RM (2014) Brain PET in the diagnosis of Alzheimer's disease. Clin Nucl Med 39: e413-422. [crossref]
- Albin RL, Koeppe RA, Burke JF, Giordani B, Kilbourn MR, et al. (2010) Comparing Fludeoxyglucose F18–PET Assessment of Regional Cerebral Glucose Metabolism and [11C] Dihydrotetrabenazine–PET in Evaluation of Early Dementia and Mild Cognitive Impairment. Archives of neurology 67: 440-446. [crossref]
- Ruan Q, D’Onofrio G, Sancarlo D, Bao Z, Greco A, et al. (2016) Potential neuroimaging biomarkers of pathologic brain changes in Mild Cognitive Impairment and Alzheimer’s disease: a systematic review. BMC geriatrics 16: 1. [crossref]
- Mosconi L, Berti V, Glodzik L, Pupi A, De Santi S, et al. (2010) Pre-clinical detection of Alzheimer's disease using FDG-PET, with or without amyloid imaging. J Alzheimers Dis 20: 843-854. [crossref]
- Cohen AD, Klunk WE (2014) Early detection of Alzheimer's disease using PiB and FDG PET. Neurobiol Dis 72 Pt A: 117-122. [crossref]
- Risacher SL, Saykin A J (2013) Neuroimaging biomarkers of neurodegenerative diseases and dementia. Semin Neurol 33: 386-416. [crossref]
- Ruan Q, D’Onofrio G, Sancarlo D, Bao Z, Greco A, et al. (2016) Potential neuroimaging biomarkers of pathologic brain changes in Mild Cognitive Impairment and Alzheimer’s disease: a systematic review. BMC geriatrics 16: 1. [crossref]
- Luckhaus C, Cohnen M, Flüb MO, Jänner M, Grass-Kapanke B, et al. (2010) The relation of regional cerebral perfusion and atrophy in mild cognitive impairment (MCI) and early Alzheimer's dementia. Psychiatry Research: Neuroimaging 183: 44-51. [crossref]
- Hauser T, Schönknecht P, Thomann PA, Gerigk L, Schröder J, et al. (2013) Regional cerebral perfusion alterations in patients with mild cognitive impairment and Alzheimer disease using dynamic susceptibility contrast MRI. Academic radiology 20: 705-711. [crossref]
- Borroni B, Anchisi D, Paghera B, Vicini B, Kerrouche N, et al. (2006) Combined 99mTc-ECD SPECT and neuropsychological studies in MCI for the assessment of conversion to AD. Neurobiol Aging 27: 24-31. [crossref]
- Chao LL, Buckley ST, Kornak J, Schuff N, Madison C, et al. (2010) ASL perfusion MRI predicts cognitive decline and conversion from MCI to dementia. Alzheimer disease and associated disorders 24: 19. [crossref]
- Caroli A, Testa C, Geroldi C, Nobili F, Barnden LR, et al. (2007) Cerebral perfusion correlates of conversion to Alzheimer's disease in amnestic mild cognitive impairment. Journal of neurology 254: 1698-1707. [crossref]
- Shokouhi S, Campbell D, Brill AB, Gwirtsman HE (2016) Longitudinal Positron Emission Tomography in Preventive Alzheimer's Disease Drug Trials, Critical Barriers from Imaging Science Perspective. Brain Pathology 26: 664-671. [crossref]
- Li Y, Gilmore JH, Shen D, Styner M, Lin W, et al. (2013) Multiscale adaptive generalized estimating equations for longitudinal neuroimaging data. NeuroImage 72: 91-105. [crossref]
- Batmanghelich NK, Dalca AV, Sabuncu MR, Golland P (2013) Joint modeling of imaging and genetics. In International Conference on Information Processing in Medical Imaging. Springer Berlin Heidelberg 766-777. [crossref]
- Kong D, Giovanello KS, Wang Y, Lin W, et al. (2015) Predicting Alzheimer's Disease Using Combined Imaging-Whole Genome SNP Data. J Alzheimers Dis 46: 695-702. [crossref]
- Harrison TM, Bookheimer SY (2016) Neuroimaging genetic risk for Alzheimer’s disease in preclinical individuals: from candidate genes to polygenic approaches. Biol Psychiatry Cogn Neurosci Neuroimaging 1: 14-23. [crossref]
- Wang H, Nie F, Huang H, Yan J, Kim S, et al. (2012) From phenotype to genotype: an association study of longitudinal phenotypic markers to Alzheimer's disease relevant SNPs. Bioinformatics 28: i619-i625. [crossref]
- Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, et al. (2014) Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain imaging and behavior 8: 183-207. [crossref]
- Tan L1, Yu JT, Zhang W, Wu ZC, Zhang Q, et al. (2013) Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement 9: 546-553. [crossref]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics 41: 1088-1093. [crossref]
- Shen L, Jia J (2016) An Overview of Genome-Wide Association Studies in Alzheimer's Disease. Neurosci Bull 32: 183-190. [crossref]
- Yang X, Li J (2016) Impact of PICALM and CLU on hippocampal degeneration. Hum Brain Mapp 37: 2419-2430. [crossref]
- Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, et al. (2011) Hippocampal function in healthy carriers of the CLU Alzheimer's disease risk variant. J Neurosci 31: 18180-18184. [crossref]
- DiBattista AM, Stevens BW, Rebeck GW, Green AE (2014) Two Alzheimer’s disease risk genes increase entorhinal cortex volume in young adults. Frontiers in human neuroscience 8. [crossref]
- Schjeide BMM, Schnack C, Lambert JC, Lill CM, Kirchheiner J, et al. (2011) The role of clusterin, complement receptor 1, and phosphatidylinositol binding clathrin assembly protein in Alzheimer disease risk and cerebrospinal fluid biomarker levels. Archives of general psychiatry 68: 207-213. [crossref]
- Lambert JC1, Zelenika D, Hiltunen M, Chouraki V, Combarros O, et al. (2011) Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging 32: 756. [crossref]
- Schott JM; ADNI Investigators (2012) Using CSF biomarkers to replicate genetic associations in Alzheimer's disease. Neurobiol Aging 33: 1486. [crossref]
- Voineskos AN, Felsky D, Wheeler AL, Rotenberg DJ, Levesque M, et al. (2015) Limited Evidence for Association of Genome-Wide Schizophrenia Risk Variants on Cortical Neuroimaging Phenotypes. Schizophrenia bulletin sbv180. [crossref]
- Louwersheimer E, Ramirez A, Cruchaga C, Becker T, Kornhuber J, et al. (2015) Influence of genetic variants in SORL1 gene on the manifestation of Alzheimer's disease. Neurobiol Aging 36: 1605. [crossref]
- Benussi L, Ghidoni R, Pegoiani E, Moretti DV, Zanetti O, et al. (2009) Progranulin Leu271LeufsX10 is one of the most common FTLD and CBS associated mutations worldwide. Neurobiology of disease 33: 379-385. [crossref]
- Morris CM, Ballard CG, Allan L, Rowan E, Stephens S, et al. (2011) NOS3 gene rs1799983 polymorphism and incident dementia in elderly stroke survivors. Neurobiol Aging 32: 554. [crossref]
- Schuur M, Van Swieten JC, Schol-Gelok S, Ikram MA, Vernooij MW, et al. (2010) Genetic risk factors for cerebral small-vessel disease in hypertensive patients from a genetically isolated population. J Neurol Neurosurg Psychiatry 2009. [crossref]
- Inkster B, Rao AW, Ridler K, Filippini N, Whitcher B, et al. (2012) Genetic variation in GOLM1 and prefrontal cortical volume in Alzheimer's disease. Neurobiology of aging 33: 457-465. [crossref]
- Lyall DM, Royle NA, Harris SE, Bastin ME, Maniega SM, et al. (2013) Alzheimer’s disease susceptibility genes APOE and TOMM40, and hippocampal volumes in the Lothian birth cohort 1936. PloS one 8: e80513. [crossref]
- Assareh AA, Piguet O, Lye TC, Mather KA, Broe GA, et al. (2014) Association of SORL1 gene variants with hippocampal and cerebral atrophy and Alzheimer's disease. Curr Alzheimer Res 11: 558-563. [crossref]
- Oliveira-Filho J, Ornellas AC, Zhang CR, Oliveira LM, Araújo-Santos T, et al. (2015) COX-2 rs20417 Polymorphism Is Associated with Stroke and White Matter Disease. Journal of Stroke and Cerebrovascular Diseases 24: 1817-1822. [crossref]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, et al. (2013) The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer's & Dementia 9: e111-e194. [crossref]
- Kang JH, Korecka M, Figurski MJ, Toledo JB, Blennow K, et al. (2015) The Alzheimer's Disease Neuroimaging Initiative 2 Biomarker Core: A review of progress and plans. Alzheimers Dement 11: 772-791. [crossref]
- Johnson KA, Fox NC, Sperling RA, Klunk WE (2012) Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med 2: a006213. [crossref]
- Dukart J, Mueller K, Villringer A, Kherif F, Draganski B, et al. (2013) Relationship between imaging biomarkers, age, progression and symptom severity in Alzheimer's disease. NeuroImage 3: 84-94. [crossref]
- Palmqvist S, Zetterberg H, Mattsson N, Johansson P; Alzheimer's Disease Neuroimaging Initiative, Minthon L, et al. (2015) Detailed comparison of amyloid PET and CSF biomarkers for identifying early Alzheimer disease. Neurology 85: 1240-1249. [crossref]
- Mosconi L, Berti V, Glodzik L, Pupi A, De Santi S, et al. (2010) Pre-clinical detection of Alzheimer's disease using FDG-PET, with or without amyloid imaging. J Alzheimers Dis 20: 843-854. [crossref]
- Berti V, Polito C, Lombardi G, Ferrari C, Sorbi S, et al. (2016) Rethinking on the concept of biomarkers in preclinical Alzheimer's disease. Neurol Sci 37: 663-672. [crossref]
- Glahn DC, Paus T, Thompson PM (2007) Imaging genomics: mapping the influence of genetics on brain structure and function. Human brain mapping 28: 461-463. [crossref]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, et al. (2010) Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer's & Dementia 6: 265-273. [crossref]
- Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, et al. (2014) Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain imaging and behavior 8: 183-207. [crossref]
- Frisoni GB, Henneman WJ, Weiner MW, Scheltens P, Vellas B, et al. (2008) The pilot European Alzheimer's disease neuroimaging initiative of the European Alzheimer's disease consortium. Alzheimer's & Dementia 4: 255-264. [crossref]
- Villemagne VL, Kim SY, Rowe CC, Iwatsubo T (2014) Imago Mundi, Imago AD, Imago ADNI. Alzheimers Res Ther 6: 62. [crossref]
- Cavedo E, Babiloni C, Redolfi A, Lizio R, Chiapparini L, et al. (2012) The Italian Alzheimer's Disease Neuroimaging Initiative (IADNI). Alzheimer's & Dementia 8: P159. [crossref]
- Cavedo E, Redolfi A, Angeloni F, Babiloni C, Lizio R, et al. (2014) The Italian Alzheimer's Disease Neuroimaging Initiative (I-ADNI): validation of structural MR imaging. J Alzheimers Dis 40: 941-952. [crossref]
- Faux NG, Rembach A, Wiley J, Ellis KA, Ames D, et al. (2014) An anemia of Alzheimer's disease. Mol Psychiatry 19: 1227-1234. [crossref]
- Burnham SC, Faux NG, Wilson W, Laws SM, Ames D, et al. (2014) A blood-based predictor for neocortical A? burden in Alzheimer’s disease: results from the AIBL study. Molecular psychiatry 19: 519-526. [crossref]
- Ritchie CW, Molinuevo JL, Truyen L, Satlin A, Van der Geyten S, et al. (2016) Development of interventions for the secondary prevention of Alzheimer's dementia: the European Prevention of Alzheimer's Dementia (EPAD) project. Lancet Psychiatry 3: 179-186. [crossref]
- Dobkin BH (2016) CURRENT OPINION Editorial introductions.
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, et al. (2011) The parkinson progression marker initiative (PPMI). Progress in neurobiology 95: 629-635. [crossref]
- Thompson P (2014) The Enhancing Neuroimaging Genetics Through Meta-Analysis (ENIGMA) Consortium: Investigating Brain Diseases With Imaging and Genetics in 29,000 People. 61st Annual Meeting AACAP.
- Hibar DP, ENIGMA-Consortium (2013) ENIGMA2: genome-wide scans of subcortical brain volumes in 16,125 subjects from 28 cohorts worldwide. Organization of Human Brain Mapping, Seattle, WA.
- Cortese S, Castellanos FX (2012) Neuroimaging of attention-deficit/hyperactivity disorder: current neuroscience-informed perspectives for clinicians. Current psychiatry reports 14: 568-578. [crossref]
- von Rhein D, Mennes M, van Ewijk H, Groenman AP, Zwiers MP, et al. (2015) The NeuroIMAGE study: a prospective phenotypic, cognitive, genetic and MRI study in children with attention-deficit/hyperactivity disorder. Design and descriptives. European child & adolescent psychiatry 24: 265-281. [crossref]
- Younesi E, Hofmann-Apitius M (2013) Biomarker-guided translation of brain imaging into disease pathway models. Scientific reports 3. [crossref]
- Davinelli S, Intrieri M, Russo C, Di Costanzo A, Zella D, et al. (2011) The “Alzheimer's disease signature": potential perspectives for novel biomarkers. Immunity & Ageing 8: 1. [crossref]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD (2011) Large-scale automated synthesis of human functional neuroimaging data. Nature methods 8: 665-670. [crossref]
- Bairoch AM, Apweiler R, Wu CH, Barker WC, Boeckmann B, et al. (2005) The universal protein resource (UniProt). Nucleic acids research 33: D154-9. [crossref]
- Spasic I, Ananiadou S, McNaught J, Kumar A (2005) Text mining and ontologies in biomedicine: making sense of raw text. Brief Bioinform 6: 239-251. [crossref]
- Hoehndorf R, Schofield PN, Gkoutos GV (2015) The role of ontologies in biological and biomedical research: a functional perspective. Brief Bioinform 16: 1069-1080. [crossref]
- Köhler S, Bauer S, Mungall CJ, Carletti G, Smith CL, et al. (2011) Improving ontologies by automatic reasoning and evaluation of logical definitions. BMC Bioinformatics 12: 418. [crossref]
- Smith B, Arabandi S, Brochhausen M, Calhoun M, Ciccarese P, et al. (2015) Biomedical imaging ontologies: A survey and proposal for future work. J Pathol Inform 6: 37. [crossref]
- Buckler AJ, Ouellette M, Danagoulian J, Wernsing G, Liu TT, et al. (2013) Quantitative imaging biomarker ontology (QIBO) for knowledge representation of biomedical imaging biomarkers. Journal of digital imaging 26: 630-641. [crossref]
- Lasbleiz J, Saint-Jalmes H, Duvauferrier R, Burgun A (2011) Creating a Magnetic resonance imaging ontology. Studies in health technology and informatics 169: 784. [crossref]
- Lasbleiz J, Morelli J, Schnel N, Burgun A, Duvauferrier R, et al. (2012) MRI image artifact ontology: a proposed method for improved recognition. Stud Health Technol Inform 180: 103-107. [crossref]
- Barillot C, Benali H, Dojat M, Gaignard A, Gibaud B, et al. (2006) Federating distributed and heterogeneous information sources in neuroimaging: the NeuroBase project. In HealthGrid 6: 3-13. [crossref]
- Gibaud B, Kassel G, Dojat M, Batrancourt B, Michel F, et al. (2011) NeuroLOG: sharing neuroimaging data using an ontology-based federated approach. In AMIA... Annual Symposium proceedings [electronic resource]/AMIA Symposium. AMIA Symposium 2011: 472-480.
- Iyappan et al., “Neuroimaging Feature Terminology (NIFT): a controlled terminology for the annotation of brain imaging features”, Manuscript in final stages of submission
- Gattermayer T (2007) SCAIView: annotation and visualization system for knowledge discovery (Doctoral dissertation, Masters thesis, Life Science Informatics at Bonn-Aachen International Center for Information Technology (B-IT), Germany).
- Müller B, Fluck J, Hofmann-Apitius M, Friedrich CM BENCHMARKING SCAIView.
- Snyder HM, Corriveau RA, Craft S, Faber JE, Greenberg SM, et al. (2015) Vascular contributions to cognitive impairment and dementia including Alzheimer's disease. Alzheimer's & Dementia 11: 710-717. [crossref]
- Nizetic D, Chen CL, Hong W, Koo EH (2015) Inter-Dependent Mechanisms Behind Cognitive Dysfunction, Vascular Biology and Alzheimer's Dementia in Down Syndrome: Multi-Faceted Roles of APP. Frontiers in behavioral neuroscience 9. [crossref]
- Aliev G, Obrenovich M E, Smith MA, Perry G (2003) Hypoperfusion, mitochondria failure, oxidative stress, and Alzheimer disease. BioMed Research International 2003: 162-163. [crossref]
- Attems J, Jellinger KA (2014) The overlap between vascular disease and Alzheimer's disease--lessons from pathology. BMC Med 12: 206. [crossref]
- O Akinyemi R, B Mukaetova-Ladinska E, Attems J, Ihara M, N Kalaria R (2013) Vascular risk factors and neurodegeneration in ageing related dementias: Alzheimer’s disease and vascular dementia. Current Alzheimer Research 10: 642-653. [crossref]
- Snyder HM, Corriveau RA, Craft S, Faber JE, Greenberg SM, et al. (2015) Vascular contributions to cognitive impairment and dementia including Alzheimer's disease. Alzheimer's & Dementia 11: 710-717. [crossref]
- Iadecola C (2016) Vascular and Metabolic Factors in Alzheimer's Disease and Related Dementias: Introduction. Cell Mol Neurobiol 36: 151-154. [crossref]
- Zhang N, Gordon ML, Goldberg TE (2016) Cerebral blood flow measured by arterial spin labeling MRI at resting state in normal aging and Alzheimer’s disease. Neuroscience & Biobehavioral Reviews 72: 168-175. [crossref]
- Valotassiou V, Angelidis G, Papatriantafyllou J, Sifakis N, Georgoulias P (2011) The Clinical Use of SPECT and PET Molecular Imaging in Alzheimer's Disease. INTECH Open Access Publisher. [crossref]
- Bergmann H, Sinzinger H (2013) Radioactive Isotopes in Clinical Medicine and Research. Birkhäuser. [crossref]
- Abe T, Usui A (2014) [Cerebral blood flow monitoring]. Kyobu geka. The Japanese journal of thoracic surgery 67: 623-629.
- Promjunyakul NO, Lahna DL, Kaye JA, Dodge HH, Erten-Lyons D, et al. (2016) Comparison of cerebral blood flow and structural penumbras in relation to white matter hyperintensities: A multi-modal magnetic resonance imaging study. Journal of Cerebral Blood Flow & Metabolism 39: 1528-1536. [crossref]
- Wei X, Yan R, Chen Z, Weng R, Liu X, et al. (2016) Combined Diffusion Tensor Imaging and Arterial Spin Labeling as Markers of Early Parkinson’s disease. Scientific Reports 6. [crossref]
- Julien C, Tremblay C, Émond V, Lebbadi M, Salem Jr N, et al. (2009) SIRT1 decrease parallels the accumulation of tau in Alzheimer disease. Journal of neuropathology and experimental neurology 68: 48. [crossref]
- Maiese K, Chong ZZ, Shang YC, Wang S (2011) Translating cell survival and cell longevity into treatment strategies with SIRT1. Rom J Morphol Embryol 52: 1173-1185. [crossref]
- Chong ZZ, Shang YC, Wang S, Maiese K (2012) SIRT1: new avenues of discovery for disorders of oxidative stress. Expert Opin Ther Targets 16: 167-178. [crossref]
- Kumar R, Chaterjee P, Sharma PK, Singh AK, Gupta A, et al. (2013) Sirtuin1: a promising serum protein marker for early detection of Alzheimer's disease. PLoS One 8: e61560. [crossref]
- Hattori Y, Okamoto Y, Nagatsuka K, Takahashi R, Kalaria RN, et al. (2015) SIRT1 attenuates severe ischemic damage by preserving cerebral blood flow. Neuroreport 26: 113-117. [crossref]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, et al. (2005) SIRT1 protects against microglia-dependent amyloid-β toxicity through inhibiting NF-ӄB signaling. Journal of Biological Chemistry 280: 40364-40374. [crossref]
- Tang BL, Chua CE (2008) SIRT1 and neuronal diseases. Mol Aspects Med 29: 187-200. [crossref]
- Thakur N, Blanc PD, Julian LJ, Yelin EH, Katz PP, et al. (2010) COPD and cognitive impairment: the role of hypoxemia and oxygen therapy. International journal of chronic obstructive pulmonary disease 5: 263. [crossref]
- Petegnief V, Planas AM (2013) SIRT1 regulation modulates stroke outcome. Transl Stroke Res 4: 663-671. [crossref]
- Donmez G, Wang D, Cohen DE, Guarente L (2010) SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142: 320-332. [crossref]
- Ferguson D, Koo JW, Feng J, Heller E, Rabkin J, et al. (2013) Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. J Neurosci 33: 16088-16098. [crossref]
- Herskovits AZ, Guarente L (2013) Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res 23: 746-758. [crossref]
- Groot AJ, Vooijs MA (2012) The role of Adams in Notch signaling. In Notch Signaling in Embryology and Cancer Springer US 15: 36.
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, et al. (2010) ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. The EMBO journal 29: 3020-3032. [crossref]
- Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, et al. (2013) ADAM10 missense mutations potentiate β-amyloid accumulation by impairing prodomain chaperone function. Neuron 80: 385-401. [crossref]
- Kim M, Suh J, Romano D, Truong MH, Mullin K, et al. (2009) Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Human molecular genetics 18: 3987-3996. [crossref]
- Hosoi T, Nomura J, Ozawa K, Nishi A, Nomura Y (2015) Possible involvement of endoplasmic reticulum stress in the pathogenesis of Alzheimer’s disease. Endoplasmic Reticulum Stress in Diseases 2. [crossref]
- Giri M, Zhang M, Lü Y (2016) Genes associated with Alzheimer's disease: an overview and current status. Clin Interv Aging 11: 665-681. [crossref]
- Darreh-Shori T, Forsberg A, Modiri N, Andreasen N, Blennow K, et al. (2011) Differential levels of apolipoprotein E and butyrylcholinesterase show strong association with pathological signs of Alzheimer's disease in the brain in vivo. Neurobiology of aging 32: 2320-e15. [crossref]
- Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, et al. (2014) APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry 19: 351-357. [crossref]
- Vijayaraghavan S, Karami A, Aeinehband S, Behbahani H, Grandien A, et al. (2013) Regulated extracellular choline acetyltransferase activity—the plausible missing link of the distant action of acetylcholine in the cholinergic anti-inflammatory pathway. PloS one 8: e65936. [crossref]
- Darreh-Shori T, Forsberg A, Modiri N, Andreasen N, Blennow K, et al. (2011) Differential levels of apolipoprotein E and butyrylcholinesterase show strong association with pathological signs of Alzheimer's disease in the brain in vivo. Neurobiol Aging 32: 2320. [crossref]
- Ji H, Dai D, Wang Y, Jiang D, Zhou X, et al. (2015) Association of BDNF and BCHE with Alzheimer's disease: Meta-analysis based on 56 genetic case-control studies of 12,563 cases and 12,622 controls. Experimental and therapeutic medicine 9: 1831-1840. [crossref]
- Reid GA, Darvesh S (2015) Butyrylcholinesterase-knockout reduces brain deposition of fibrillar β-amyloid in an Alzheimer mouse model. Neuroscience 298: 424-435. [crossref]
- Manning EN, Barnes J, Cash DM, Bartlett JW, Leung KK, et al. (2014) APOEɛ4 is associated with disproportionate progressive hippocampal atrophy in AD. PloS one 9: e97608. [crossref]
- Li B, Shi J, Gutman BA, Baxter LC, Thompson PM, et al. (2016) Influence of APOE genotype on hippocampal atrophy over time-An N= 1925 surface-based ADNI study. PloS one 11: e0152901. [crossref]
- Cherbuin N, Sargent-Cox K, Easteal S, Sachdev P, Anstey KJ (2015) Hippocampal atrophy is associated with subjective memory decline: The PATH Through Life study. Am J Geriatr Psychiatry 23: 446-455. [crossref]
- Parikh I, Fardo DW, Estus S (2014) Genetics of PICALM expression and Alzheimer's disease. PLoS One 9: e91242. [crossref]
- Ramanathan A, Nelson AR, Sagare AP, Zlokovic BV (2015) Impaired vascular-mediated clearance of brain amyloid beta in Alzheimer’s disease: the role, regulation and restoration of LRP1. Frontiers in aging neuroscience 7. [crossref]
- Thomas RS, Henson A, Gerrish A, Jones L, Williams J, et al. (2016) Decreasing the expression of PICALM reduces endocytosis and the activity of β-secretase: implications for Alzheimer’s disease. BMC neuroscience 17: 50. [crossref]
- András IE, Toborek M (2013) Amyloid beta accumulation in HIV-1-infected brain: The role of the blood brain barrier. IUBMB Life 65: 43-49. [crossref]
- Zlokovic BV, Deane R, Sagare AP, Bell RD, Winkler EA (2010) Low-density lipoprotein receptor-related protein-1: a serial clearance homeostatic mechanism controlling Alzheimer’s amyloid β-peptide elimination from the brain. Journal of neurochemistry 115: 1077-1089. [crossref]
- Sagare AP, Deane R, Zlokovic BV (2012). Low-density lipoprotein receptor-related protein 1: a physiological A? homeostatic mechanism with multiple therapeutic opportunities. Pharmacology & therapeutics 136: 94-105. [crossref]
- Laifenfeld D, Patzek LJ, McPhie DL, Chen Y, Levites Y, et al. (2007) Rab5 mediates an amyloid precursor protein signaling pathway that leads to apoptosis. The Journal of neuroscience 27: 7141-7153. [crossref]
- Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, et al. (2015) Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat Neurosci 18: 978-987. [crossref]
- Sagare AP, Bell RD, Zlokovic BV (2013) Neurovascular defects and faulty amyloid-β vascular clearance in Alzheimer's disease. J Alzheimers Dis 33 Suppl 1: S87-100. [crossref]
- Winkler EA, Nishida Y, Sagare AP, Rege SV, Bell RD, et al. (2015) GLUT1 reductions exacerbate Alzheimer's disease vasculo-neuronal dysfunction and degeneration. Nature neuroscience 18: 521-530. [crossref]
- Lyros E, Bakogiannis C, Liu Y, Fassbender K (2014) Molecular links between endothelial dysfunction and neurodegeneration in Alzheimer's disease. Current Alzheimer Research 11: 18-26. [crossref]
- Redzic Z (2011) Molecular biology of the blood-brain and the blood-cerebrospinal fluid barriers: similarities and differences. Fluids and barriers of the CNS 8: 1. [crossref]
- De Giorgis V, Veggiotti P (2013) GLUT1 deficiency syndrome 2013: current state of the art. Seizure 22: 803-811. [crossref]
- Shah K, DeSilva S, Abbruscato T (2012) The role of glucose transporters in brain disease: diabetes and Alzheimer’s disease. International journal of molecular sciences 13: 12629-12655.
- Duran-Aniotz C, Hetz C (2016) Glucose Metabolism: A Sweet Relief of Alzheimer's Disease. Curr Biol 26: R806-809. [crossref]





