Abstract
Background: In routine clinical laboratory diagnosis, characterizing a Mycobacterial isolate and its drug susceptibility is a tedious process, largely owing to its slow growth. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a revolutionary technique which can be applied for the routine identification of Mycobacteria.
Methods: In this prospective and blinded study, we attempted to characterize 100 clinical isolates of Mycobacteria using the MALDI-TOF and evaluate its utility as a useful tool for rapid identification of clinical Mycobacterial isolates. Fresh cultures (less than 7 days) of 100 (characterized to genus level) Mycobacterial isolates, grown either on Lowenstein Jensen slants or in the Bact/Alert MP bottles, were used. The isolates were initially disrupted with ethanol and silica beads. Acetonitrile & formamide acid extraction procedure was carried out and the supernatant was spotted on the target plate and Cinnamic acid was added. The plates were subjected to identification by MALDI-TOF MS. The spectra were acquired manually and recorded in the linear positive mode at a laser frequency of 20 Hz within a mass range from 2000 to 20,000 Da. The results of identification by MALDI were compared with the results from conventional techniques.
Results: Among the 100 isolates, 80 were identified as Mycobacterium tuberculosis (M.tb) / bovis complex and 20 were non TB Mycobacteria (NTMs). There was a 100% correlation between the conventional and MALDI TOF results. The latter could identify the NTMs to the species level.
Conclusion: This study showed that the MALDI-TOF MS was found to be an accurate, rapid and robust system for identification and differentiation of Mycobacteria. This innovative approach holds promise as an alternate and competent tool in mycobacteriology and helps in initiation of timely and appropriate therapeutic intervention.
Keywords
mycobacterium tuberculosis (M.tb); identification; Non tuberculous Mycobacteria (NTM)
Introduction
Traditional methods for identifying Mycobacterial species are based on phenotypic traits, biochemical and molecular-based methods which include PCR and DNA sequencing of a variety of target genes. Though these methods are accurate, they are resource and time-consuming [1]. Reducing the time associated with the identification of the Mycobacterial isolates can be achieved by new and cost-effective methods of detection [2,3].
Recently, several studies have provided the proof-of-concept that MALDI-TOF MS is a rapid, cost- effective and competent alternate method for the precise identification and differentiation of Mycobacterial species directly from cultures [4-8]. Compared to the traditional biochemical and phenotypic methods or molecular assays, the MALDI TOF method is user friendly with a rapid turn - around time of one hour [9-14].
In this prospective study, the prime aim was to use VITEK® MS - a MALDI-TOF mass spectrometer (bioMerieux, France) to characterize 100 clinical Mycobacterial isolates. The database available on this platform was implemented to generate reliable identification through recognizing the unique protein fingerprints of the Mycobacterial isolates.
Methods
A total of 100 clinical isolates of Mycobacteria from various clinical specimen (Pulmonary and Extra pulmonary) submitted for Mycobacterial cultures to the Department of Microbiology, Nizam’s Institute of Medical Sciences, Hyderabad, Telangana State, India., were included in the study. The isolates were identified as M.tb or NTMs (genus level) by conventional methods.
Fresh cultures of these 100 Mycobacterial isolates, in their exponential growth phase (14 -21 days old), on BBL™ Lowenstein-Jensen Medium Slants, BD, were used for the analysis. Standard strain of M.tb, H37RV was also included and processed similarly as the clinical isolates.
All the procedures were performed in a biosafety cabinet using all safety and aseptic precautions. The method of extraction recommended and standardized by bioMerieux (France) for the VITEK® MS was used [1]
Briefly, one µl loopful of the mycobacterial biomass was mixed in a 1.5-ml sterile screw-top micro-centrifuge tube containing 900 µl of 70% ethanol, to obtain a turbid suspension and kep at 37C for 15 mts for inactivation of the organisms. To this suspension, 200 µl of 0.5-mm sterile silica beads were added and the tubes were vortexed for 15 mts in a horizontal position using a Vortex-Genie 2 with a 24-tube adaptor at maximum speed (Scientific Industries Inc, USA) with MoBio Vortex Adaptor) (MoBio Laboratories, Inc.,USA) to dissociate the Mycobacterial cell walls. Three rounds of dissociation were done with an interval of 10 mts for cooling, between each round. The tubes were then incubated at 37 0C for 10 min for the beads to settle down. The supernatant was then transferred, with a sterile pipette, to a new sterile 1.5-ml snap-top micro-centrifuge tube, taking care that no beads are transferred. They were then centrifuged at 10,000 g for 2 min. The supernatant was decanted, and the excess removed with a narrow-gauge pipette. The pellets were air dried with the tube open to ensure complete evaporation of the ethanol. To the dried pellets, 20 µl of acetonitrile was added and vortexed, followed by addition of 20 µL of 70% formamide and vortexing. The tubes were centrifuged at 2000 g for 10 minutes and 1𝜇L of the supernatant was spotted in triplicate on the steel target plate of the VITEK® MS. The spots were allowed to dry at room temperature. Finally, 1𝜇L of CHCA matrix solution (𝛼-cyano- hydroxycinnamic acid) was added to each spot and left to dry before they were loaded on to the VITEK® MS instrument. A quality control organism, E.coli (ATCC 8739) was included in every plate assayed which served as an assay control as well as a negative control.
As recommended by bioMerieux, the Escherichia coli ATCC 8739 strain, used as a calibrator and internal ID control, was inoculated on the calibration spots of each acquisition groups (small spot in the middle of each acquisition group). Each bacterial isolate had been tested with a unique deposit.
A minimum of 100 laser shots per sample were used to generate each spectrum.
Analysis
The samples were analyzed in triplicate using a Microflex LT MALDI-TOF MS instrument. The spectra were acquired manually and recorded in the linear positive mode at a laser frequency of 20 Hz within a mass range from 2000 to 20,000 Da. Parameter settings for Microflex instrument were ion source 1 at 20 kV, ion source 2 at 18.5 kV, lens at 8.5 kV, pulsed ion extraction of 250 ns and no gating.
A comparative analysis of the results of identification of the 100 clinical mycobacterial isolates using the MALDI-TOF MS and conventional techniques was carried out.
Results
All the 100 Mycobacterial isolates yielded a visible protein profile with an identification score of 95-99.9% confidence. Eighty of the isolates were identified as M. tb / M.bovis. Of these, 78 M.tb isolates had a score of 50:50. The scores were equally distributed between the 2 Mycobacterial species (M.tb & M.bovis). The remaining 2 isolates, out of the 80, had a score of 25:25 but were interpreted as Mtb / M.bovis by the data base.
The remaining 20 isolates were interpreted as non-TB Mycobacteria. Of these, 13 were identified as M.fortuitium with scores of 99.9%. Table shows the distribution of the NTMs and their scores.
The isolates were differentiated by mass spectral (MS) profiles that were distinct among each of the Mycobacterial species (Figure 1). As per the MS profiles for M.tb, signals between 10,000 to 12500 m/z were very distinct and were present in each of the 80 M.tb isolates and the standard H37RV.
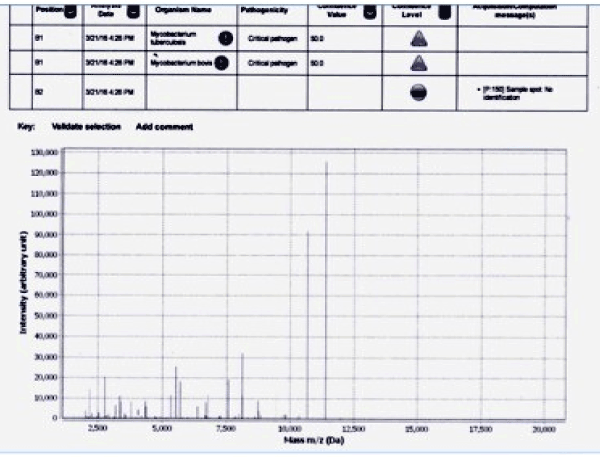
Figure 1. MS profile of Mycobacterium tuberculosis complex
The MS profiles of the NTM`s showed signals below 6000m/z and were very distinctly different from those of M.tb (Figure 2). The different MS profiles obtained are shown in Figure 2. M.avium and M.africanum had very close peaks below 4000 m/z while M.flavescens showed very few signals but with a peak at 5750 m/z. The standard strains of the NTMs also showed similar species-specific profiles.
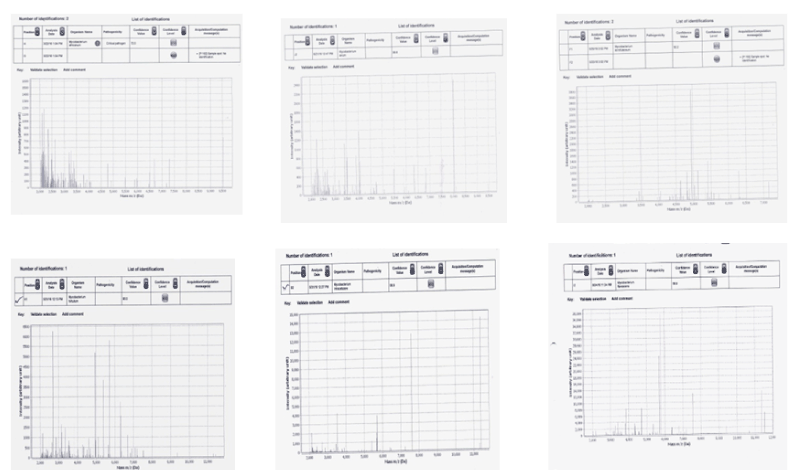
Figure 2. MS profiles of atypical mycobacteria
1-Mycobacterium africanum
2-Mycobacterium avium
3-Mycobacterium scrofulaceum
4-Mycobacterium fortuitum
5-Mycobacterium intracellulare
6-Mycobacterium flavescans
In all cases, the negative control (E. coli) yielded visible peaks that were very distinct from those of Mycobacteria.
Discussion
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) has emerged over the last few years as a novel tool for rapid and reliable identification of microorganisms by analysis of protein profiles from either disrupted cells or intact bacterial cells. The accurate identifications and reduced turnaround times and cost, are facilitating an appropriate management of the infectious diseases [9,15,16].
MALDI-TOF MS has been recently used to characterize mycobacteria [5,8,10]. In India, the technology has been successfully adapted for the rapid identification of Mycobacteria compared to conventional phenotypic identification methods [14,18]. The method can analyze Mycobacterial isolates rapidly within minutes and thereby facilitate high throughput outcome. The simple extraction procedure, low running cost and the non-requirement of high technical expertise provide MALDI-TOF MS an edge over other methods for identification [19,14]. It has been observed that MALDI-TOF MS generates less waste than other methods that are based on molecular and biochemical tests that use many disposable materials [1].
Sample preparation techniques define the quality and composition of protein expression profiles obtained on MALDI TOF [11,20,].
Several studies have shown that the sample preparation techniques define the quality and composition of protein expression profiles obtained on MALDI TOF. An original protocol for the MALDI-TOF MS identification of heat-inactivated Mycobacteria after dissociation in Tween-20, mechanical breaking of the cell wall and protein extraction with formic acid and acetonitrile was described by El et al. [11]. By applying this protocol to as few as 105 colony-forming units of reference isolates of M.tb, M. avium, and 20 other Mycobacterium species, they obtained species-specific mass spectra for the creation of a local database. Using this database, their protocol enabled the identification of 87 M.tb, 25 M.avium and 12 non-tuberculosis clinical isolates within 2.5 hours and with identification scores ≥2. Further analysis of the profiles showed that certain signals ranging from 3 to 10 kDa were identified as “species-specific biomarkers” at m/z 5519, 5700, 7100, 8336, 9270, 10,662, and 11,376 [5].
In the present study, the biomerieux’s method of extraction was used as the analysis was on Vitek MS [1]. Within the observed mass range, a few unique signals were conserved across all the 80 M.tb/ M.bovis isolates similar to the studies conducted by Hettick et al. [12] El Khéchine et al. [11] and Saleeb et al. [13] on different M. tb isolates, thereby highlighting these signals to be potential “species-specific biomarkers.” Furthermore, it was observed that there was a difference in the peak height ratio across the spectra within the species among the NTMs (Figure 2). This could be attributed to the differential expression of the same protein among different species.
Conclusion
As per this study the MALDI-TOF MS provided an accurate, rapid identification and differentiation of Mycobacteria. The extraction process was simple without issue of contamination and biohazard risks to the technical personnel.
Ethics approval
The protocol of the experiments was reviewed and approved by the Ethics Committee of NIMS (reference no. EC/NIMS/1755/2016). Availability of data and materials- All data and materials are available.
Competing interests
2021 Copyright OAT. All rights reserv
The authors declare that they have no competing interests.
Declaration
We thank GYD Diagnostics Pvt ltd for permitting us to use their VITEK-MS.
Authors contribution
LV & LA designed the experiments and drafted the manuscript. LA performed the experiments. LV and LA helped draft the manuscript. LV, LA, SD and VY equally participated in the study design and coordinated the research work. All authors read and approved the final manuscript.
Funding
We are grateful to GYD Diagnostics Pvt Ltd for their support in terms of providing reagents and target slides necessary for the study.
References
- Llasat BJM, Kamboj K, Pancholi P (2013) Identification of Mycobacteria from Solid and Liquid Media by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry in the Clinical Laboratory. J Clin Microbiol 51: 2875-2879
- Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O, et al. (2011) (Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6: e16424.
- Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G, et al. (2010) Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol 48:1549-1554.
- Mather CA, Rivera SF, Wu SMB (2014) Comparison of the Bruker Biotyper and Vitek MS Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Systems for Identification of Mycobacteria Using Simplified Protein Extraction Protocols. J Clin Microbiol 52: 130 -138
- Cherkaoui A, Hibbs B, Emonet S, Tangomo S, Girard M, et al. (2010) Comparison of two matrix-assisted laser desorption ionization-time offlight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol 48: 1169-1175
- Dhiman N, Hall L, Wohlfiel SL, Buckwalter SP, Wengenack NL (2011) Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 49: 1614-1616 [Crossref]
- El Khéchine A, Couderc C, Flaudrops C, Raoult D, Drancourt M (2011) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6: e24720. [Crossref]
- Gaillot O, Blondiaux N, Loiez C, Wallet F, Lemaître N, et al. (2011) Cost-effectiveness of switch to matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine bacterial identification. J Clin Microbiol 49: 4412. [Crossref]
- Hettick JM, Kashon ML, Slaven JE, Ma Y, Simpson JP, et al. (2006) Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistical analysis. Proteomics 6: 6416-6425. [Crossref]
- Lotz A, Ferroni A, Beretti JL, Dauphin B, Carbonnelle E, et al. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 48: 4481-4486
- Martiny D, Busson L, Wybo I, Haj RAE, Dediste A [2012] Comparison of the Microflex LT and Vitek(R) MS systems for the routine identification of bacteria by Matrix-Assisted Laser Desorption-Ionization Time-Of-Flight Mass Spectrometry. J. Clin. Microbiol 50: 1313-25.
- Panda A, Kurapati S, Samantaray JC, Myneedu VP, Verma A, et al. (2013) Rapid identification of clinical mycobacterial isolates by protein profiling using matrix assisted laser desorption ionization-time of flight mass spectrometry. Indian J Med Microbiol 31: 117-122. [Crossref]
- Park JS, Choi SH, Hwang SM, Hong YJ, Kim TS, et al. (2016) The impact of protein extraction protocols on the performance of currently available MALDI-TOF mass spectrometry for identification of mycobacterial clinical isolates cultured in liquid media. Clin. Chim. Acta 460: 190-195
- Ravva SV, Harden LA and Sarreal CZ (2017) Characterization and Differentiation of Mycobacterium avium subsp. paratuberculosis from Other Mycobacteria Using Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Front. Cell. Infect. Microbiol 7: 1-297
- Shitikov E, Ilina E, Chernousova L, Borovskaya A, Rukin I, et al. (2011) Mass spectrometry based methods for the discrimination and typing of mycobacteria. Infect. Genet. Evol.
- Saleeb PG, Drake SK, Murray PR, Zelazny AM (2011) Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry? J Clin Microbiol 49:1790-1794
- AzamlA A, Alki A (2016) Comparison of MALDI-TOF MS, nucleic acid hybridization and the MPT64 immunochromatographic test for the identification of M. tuberculosis and non-tuberculosis Mycobacterium species. New Microbiol 39: 259-263.
- Seng P, Drancourt M, Gouriet F, Scola BL, Fournier PE, et al. (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis 49: 543-551.
- Wilen CB, McMullen AR, Burnham C-AD (2015) Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant mycobacteria by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53: 2308-2315
- Zingue D, Flaudrops C, Drancourt M (2016) Direct matrix- assisted laser desorption ionisation time-of-flight mass spectrometry identification of mycobacteria from colonies. Eur. J. Clin. Microbiol. Infect. Dis 35: 1983-1987.


