The aim of this study is to find the relationship between HIV-1 activity and chemical structure for 2-Pyridinone derivatives by using the Electron-Topological Method (ETM). Data for ETM were obtained quantum mechanical calculations. Quantum chemical calculations were performed after the conformational analysis. By using the data obtained from quantum chemical calculation results ETM were perfomed and pharmacophere and anti-pharmacophere fragments for the HIV-1-specific Reverse Transcriptase inhibitors were explained. Conformational analysis and quantum-chemical calculations of 2-pyridinone derivatives were carried out by using B3LYP method with basis set of the 6-311G(d,p) in order to determine molecular properties. The descriptors of HOMO, LUMO, HOMO-LUMO energy gap, chemical hardness, chemical softness, electro-negativity, chemical potential, dipole moment etc. were calculated and tabulated in order to employed in statistical analyses that are Linear Discriminant Analysis (LDA) and Artificial Neural Networks (ANNs). By doing so, the linear and non-linear sections of data structure are investigated and their corresponding descriptors having impact on dependent variable has been found.
We see from the fragment properties atoms found in benzoxazole groups give rise to activity of the molecules, and atoms in the naphthyl groups causes breaking the activity.
electron topological method, HIV-1, pyridinone derivatives, structure-activity relationships, B3LYP
The identification of human immunodeficiency virus type-1 (HIV-1) as a causal agent of acquired immunodeficiency syndrome (AIDS) has led to intense research efforts to find effective therapies for this disease. Although there are inhibitors targeting various stages of the life cycle of human immunodeficiency virus (HIV), but only reverse transcriptase (RT) and protease inhibitors are currently being used to treat this disease [1-3].
Reverse transcription means the passage of an RNA genome to a double-stranded DNA molecule. This process was first reported Temin and Mizutani [4] for RNA tumor viruses. Since all previously known transfers of genetic information are made from DNA to RNA, the synthesis of retro viral DNA from an RNA genome has been termed as "reverse" transcription, ie reverse transcription. All viruses that are dependent on their proliferation in the reverse transcript are grouped in the Retroviridae family.
Reverse transcriptase (RT) is an important target for AIDS antiviral drug therapies. This enzyme is blocked by nucleoside analogues that function as chain terminators during the replication of newly synthesized proviral DNA from viral RNA.
However, their usefulness has remained limited due to toxic side effects and emergence of resistant strains of HIV-1. Non-nucleoside RT inhibitors (NNRTIs) usually contain a class of potent antiviral agents that are non-competitive inhibiting and bind to a unique site on the structure of HIV-1 RT [5,6].
It was reported that TIBO (4,5,6,7-tetrahydro-5-methylimidazo[4,5,1-j,k] [1,4] benzodiazepin-2(1H)-one) derivatives inhibited HIV-1 replication. Electron-Topological Method (ETM) [7-11] were used to study structure-activity relationships for different series [7-11].
QSAR studies were performed by Garg, et al. [12] for the pyridinone analogues and suggested a positive role of hydrophobic 20 and 21 positions R-substituents. Electron-releasing R substituents that are marginal at best seem to favour activity, and there is detrimental steric effect of R substituents. The R-substituents at 22 and 23 positions have steric effects.
In this study, structure-activity relationships have been performed for 2-pyridinone series (Table 1). The series of compounds subjected to ETM calculations studies are given in Table 1 where the activity parameter IC50 is a measure of antiviral potency and refers to the molar concentration of the compound, required to reduce the concentration of HIV-1 reverse transcriptase. The ETM modelling is a method to elucidate fragments presenting activity and represents fragments breaking activity. ETM methods have been applied to drug design problems. Besides, two statistical models conducted for the determination of statistically significant descriptors for the linear and non-linear parts of the data set are Linear Discriminant Analysis (LDA) and Artificial Neural Networks (ANNs), respectively.
Table 1. A list of molecules under investigation [13,14]. a2 naphthyl instead of benzoxazole.

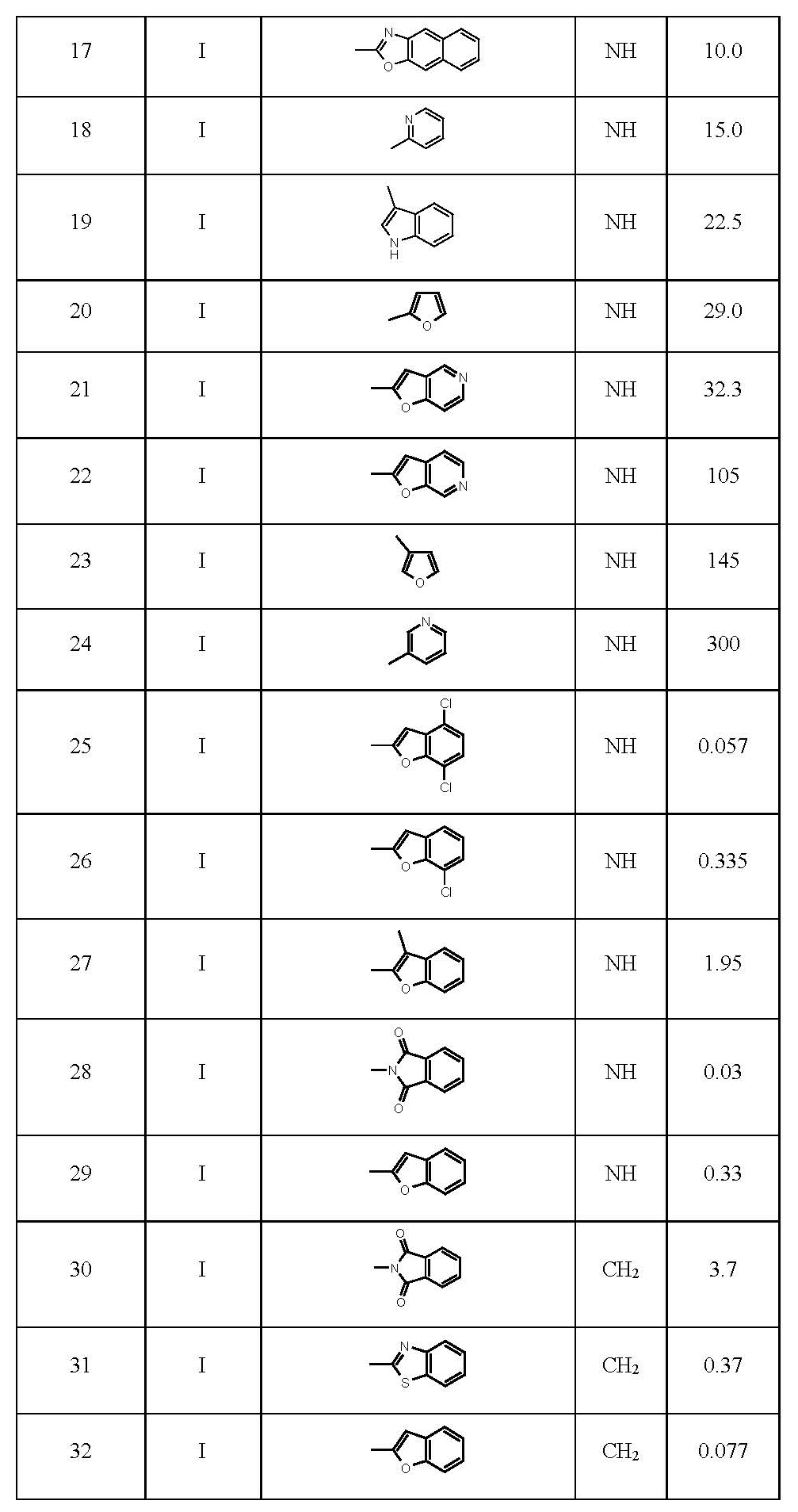

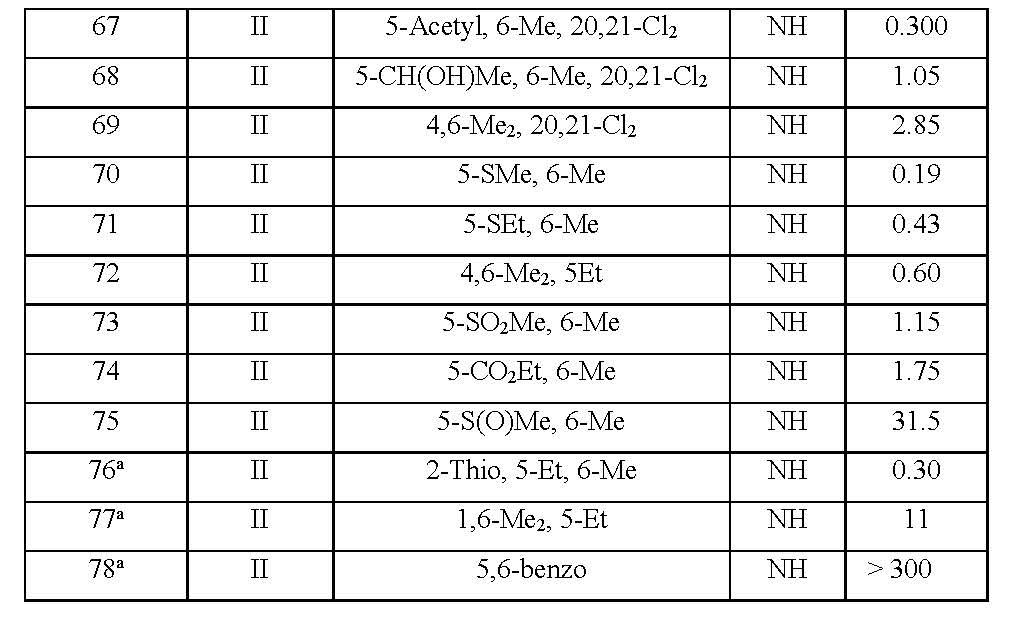
Data set on the HIV-1-Specific Reverse Transcriptase Inhibitors
2-Pyridinone derivatives whose biological activity taken from the literature [13,14] were selected to determine pharmacophore properties causing activity or inactivity. The compounds investigated were divided into two common structure as shown in Figure 1.

Figure 1. Skeletons of studied compounds.
78 Compounds in the series are divided into three groups according to their inhibition of human immunodeficiency virus type 1 reverse transcriptase as active molecules whose IC50 values between 0.0096 and 0.29 mM, low ones between 0.29 and 1.26 mM, inactive ones between 1.26 and 310 mM.
ETM [15] was applied to active compounds and inactive ones separately to reveal pharmacophore feature.
Brief description of the ET-method
The calculations under the Electronic-Topological approach are represented by a result of the following steps [16-20]:
- Conformational analysis.
- Quantum-chemical calculations.
- ETMC formation
- ETMC processing and search of the structural features of activity (pharmacophores-Ph) or inactivity (anti-pharmacophores-AP).
Computational details
In this section of study, all calculations were carried out using DFT/B3LYP method with Gaussian program [21]. Optimization of molecules was performed with 6-311G (d,p) basis set. This basis set is known as one of the basis sets that gives more accurate results in terms of the determination of electronic and geometries properties for a wide range of organic compounds. Quantum chemical parameters, the energy of the highest occupied molecular orbital (EHOMO), the energy of the lowest unoccupied molecular orbital (ELUMO), HOMO-LUMO energy gap (ΔE), dipole moment (DM), molar volume (MV), total negative charge (TNC), chemical hardness (ղ) and softness (s), electronegativity (c), chemical potential (m), global electrophilicity (ω), sum of the total negative charge (TNC) and sum of electronic and zero-point energies (SEZPE), were calculated by 6-311G (d,p) basis set of B3LYP method and discussed.
Molecular properties associated with related to the reactivity and selectivity of the compounds were estimated following the Koopmans’s theorem relating the energy of the HOMO and the LUMO. Electronegativity is estimated using the following the equation:
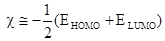 (1)
(1)
Chemical hardness (h) measures the resistance of an atom to a charge transfer [22], it is calculated by using the equation:
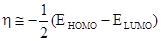 (2)
(2)
Electron polarizability, called chemical softness (s), describes the capacity of an atom or group of atoms to receive electrons [22] and is estimated by using equ. (3):
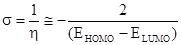 (3)
(3)
Chemical potential (m) and electronegativity (c) can be calculated with the help of the following equations [23].
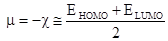 (4)
(4)
The global electrophilicity (w) is a useful reactivity descriptor that can be used to compare the electron-donating abilities of molecules [24]. Global electrophilicity index is estimated by using the electronegativity and chemical hardness parameters with below equation:
w = 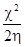 (5)
(5)
A high value of electrophilicity describes a good electrophile while a small value of electrophilicity describes a good nucleophile [25].
All the conformational and quantum-chemical data for molecules under study were obtained by the MM2P method and the semi-empirical quantum-chemical method ММ1 of molecular mechanics. We used to find out ETM-software for activity features’ selection.
These are chemically bonded and chemically unbounded atomic pairs, respectively. Every active compound in the series. As using every active compound as a template to compare with the rest of ETMCs, Molecule 60 was chosen as template compound and we found pharmacophore Ph1 being found 20 in active compounds and only one inactive ones. Ph1 include atoms in the benzoxazole ring, and C12 as seen Figure 2a. The probability of realizing Pa in this class is nearly 0.90. Ph2 was obtained as taken molecule 61 template compound. Pharmacophore 2 includes 5 atoms belonging to four of them benzoxazole ring and one is C4. The distance between C4 and C22 is 8.95 Å as seen Figure 2b. This feature was found in 21 active and 1 inactive compounds.
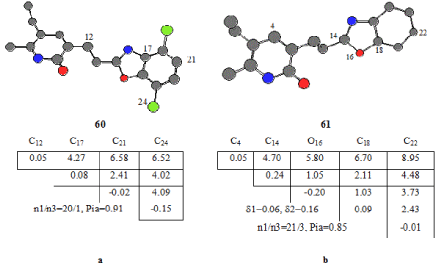
Figure 2. ETSC and corresponding structure of the pharmacophores Ph1 and Ph2 found relative to active compounds 60 and 61, respectively.
78 and 24 compounds in inactive ones were taken as template compounds to determine anti-pharmacophores. As seen from Figure 3a. AP1 includes atoms in naphthyl group and C25 in benzo pyridine ring. This feature was seen in 23 inactive compounds; the probability is 0.97. AP2 consists of four atoms being C4, C5, C11, C18. (Figure 3b).
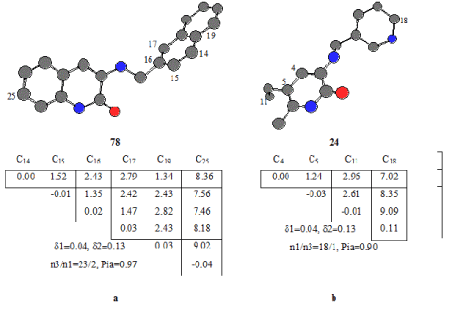
Figure 3. ETSC and corresponding structure of the anti-pharmacophores AP1 and AP2 found relative to inactive compounds 78 and 24, respectively.
(Aminomethyl)phthalimide derivatives contain benzoxazole group in Figure 4. Introduction of NH2, NO2, phenyl groups into available positions on to benzoxazole ring decreases inhibitory potency. Thus, the compound 35 is active while the other compounds are inactivated.
Figure 4. Comparison of pharmacophores.
As seen from Figure 5, both pharmacophores appear with high values of frequencies in the class of active compounds being practically absent in the class of inactive compounds. In a similar way, AP1 and AP2 are observed in maximal values for the in the class of inactive compounds while for the Ph1 and Ph2 the frequencies are almost close to zero. They are practically very low levels in the class of inactive compounds.
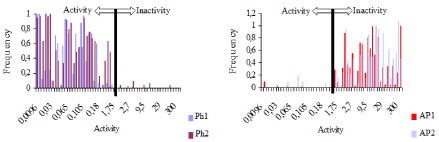
Figure 5. Frequency of the fragments’ occurrences in the compounds studied: for pharmacophores Ph1and Ph2; and for antipharmacophores AP1 and AP2.
Similarly, maximum values for frequencies of AP1 and AP2 appearances in the class of inactive compounds are observed, but the frequencies for P1 and P2 are close to zero
Molecular structure
EHOMO, ELUMO, DE, DM, MV, TNC, h, s, m, c, ω, SEZPE were calculated for 2-pyridinone derivatives (1-78) with the B3LYP/6-311G(d,p) method, as shown in Table 2.
Table 2. The calculated quantum chemical parameters for investigation compounds using B3LYP/6-311G(d,p) method.
Mol. |
EHOMO
(eV) |
ELUMO
(eV) |
∆E
(eV) |
DM
(D) |
MV
(cm3/mol) |
TNC
(e) |
h
(eV) |
s
(eV-1) |
c
(eV) |
µ
(eV) |
ω
(D2/eV) |
SEZPE
(eV) |
1 |
-5.07 |
-0.72 |
4.35 |
3.707 |
221.884 |
-3.992 |
2.173 |
0.460 |
2.894 |
-2.894 |
3.162 |
-25527.731 |
2 |
-5.32 |
-1.49 |
3.82 |
5.452 |
228.958 |
-3.333 |
1.912 |
0.523 |
3.405 |
-3.405 |
7.772 |
-25490.558 |
3 |
-5.06 |
-1.13 |
3.93 |
2.742 |
216.818 |
-3.295 |
1.965 |
0.509 |
3.097 |
-3.097 |
1.913 |
-34220.358 |
4 |
-5.19 |
-1.09 |
4.10 |
4.212 |
239.054 |
-3.071 |
2.049 |
0.488 |
3.141 |
-3.141 |
4.329 |
-25053.729 |
5 |
-5.29 |
-0.96 |
4.32 |
4.744 |
177.798 |
-3.234 |
2.162 |
0.463 |
3.125 |
-3.125 |
5.206 |
-33783.192 |
6 |
-4.81 |
-1.65 |
3.16 |
1.865 |
241.085 |
-3.274 |
1.579 |
0.633 |
3.229 |
-3.229 |
1.101 |
-25490.729 |
7 |
-5.25 |
-1.46 |
3.78 |
4.936 |
225.997 |
-3.807 |
1.892 |
0.529 |
3.355 |
-3.355 |
6.440 |
-28111.157 |
8 |
-5.28 |
-1.34 |
3.94 |
2.129 |
204.652 |
-3.521 |
1.971 |
0.507 |
3.308 |
-3.308 |
1.150 |
-25868.317 |
9 |
-5.09 |
-1.29 |
3.80 |
3.515 |
206.798 |
-3.075 |
1.900 |
0.526 |
3.190 |
-3.190 |
3.252 |
-25053.628 |
10 |
-5.04 |
-1.26 |
3.78 |
2.243 |
218.818 |
-3.463 |
1.890 |
0.529 |
3.153 |
-3.153 |
1.331 |
-27537.622 |
11 |
-5.14 |
-1.41 |
3.74 |
5.197 |
202.259 |
-4.011 |
1.868 |
0.535 |
3.277 |
-3.277 |
7.230 |
-27975.826 |
12 |
-5.23 |
-1.31 |
3.92 |
3.229 |
286.259 |
-3.570 |
1.960 |
0.510 |
3.274 |
-3.274 |
2.659 |
-29612.090 |
13 |
-5.21 |
-1.12 |
4.08 |
7.285 |
219.944 |
-3.550 |
2.042 |
0.490 |
3.166 |
-3.166 |
12.993 |
-24453.742 |
14 |
-5.16 |
-0.85 |
4.31 |
4.046 |
225.463 |
-2.890 |
2.157 |
0.464 |
3.006 |
-3.006 |
3.795 |
-20873.247 |
15 |
-5.02 |
-0.77 |
4.25 |
2.235 |
190.404 |
-3.511 |
2.125 |
0.470 |
2.896 |
-2.896 |
1.175 |
-24890.800 |
16 |
-5.02 |
-0.78 |
4.24 |
4.219 |
235.746 |
-3.579 |
2.120 |
0.472 |
2.903 |
-2.903 |
4.198 |
-25118.109 |
17 |
-5.26 |
-1.50 |
3.76 |
3.253 |
227.430 |
-3.568 |
1.881 |
0.532 |
3.380 |
-3.380 |
2.813 |
-29612.005 |
18 |
-4.84 |
-0.96 |
3.88 |
1.709 |
149.066 |
-2.999 |
1.942 |
0.515 |
2.898 |
-2.898 |
0.752 |
-21310.156 |
19 |
-4.98 |
-0.70 |
4.27 |
1.901 |
211.705 |
-3.402 |
2.137 |
0.468 |
2.842 |
-2.842 |
0.846 |
-24453.474 |
20 |
-5.12 |
-0.81 |
4.31 |
3.244 |
201.723 |
-2.931 |
2.153 |
0.465 |
2.964 |
-2.964 |
2.444 |
-20813.622 |
21 |
-5.31 |
-0.98 |
4.32 |
6.030 |
216.899 |
-3.479 |
2.162 |
0.463 |
3.143 |
-3.143 |
8.409 |
-25431.093 |
22 |
-5.32 |
-1.12 |
4.20 |
5.192 |
188.868 |
-3.248 |
2.098 |
0.477 |
3.220 |
-3.220 |
6.425 |
-25431.065 |
23 |
-5.19 |
-0.87 |
4.32 |
4.286 |
171.738 |
-2.952 |
2.159 |
0.463 |
3.026 |
-3.026 |
4.254 |
-20813.520 |
24 |
-5.31 |
-0.99 |
4.32 |
5.489 |
154.031 |
-3.102 |
2.162 |
0.463 |
3.151 |
-3.151 |
6.969 |
-21310.022 |
25 |
-5.31 |
-1.25 |
4.07 |
4.407 |
239.488 |
-3.420 |
2.034 |
0.492 |
3.281 |
-3.281 |
4.775 |
-50008.672 |
26 |
-5.23 |
-0.97 |
4.27 |
3.011 |
220.327 |
-3.373 |
2.133 |
0.469 |
3.102 |
-3.102 |
2.125 |
-37501.495 |
27 |
-5.23 |
-0.92 |
4.31 |
4.320 |
222.379 |
-3.498 |
2.157 |
0.464 |
3.072 |
-3.072 |
4.327 |
-26063.809 |
28 |
-5.18 |
-2.37 |
2.81 |
2.794 |
220.579 |
-3.931 |
1.406 |
0.711 |
3.774 |
-3.774 |
2.775 |
-28516.933 |
29 |
-5.18 |
-0.87 |
4.31 |
3.603 |
253.760 |
-3.227 |
2.156 |
0.464 |
3.026 |
-3.026 |
3.011 |
-24994.355 |
30 |
-5.80 |
-2.23 |
3.57 |
3.403 |
235.251 |
-3.987 |
1.783 |
0.561 |
4.014 |
-4.014 |
3.248 |
-28080.295 |
31 |
-5.87 |
-1.27 |
4.60 |
4.743 |
229.796 |
-3.440 |
2.299 |
0.435 |
3.568 |
-3.568 |
4.893 |
-33783.745 |
32 |
-5.77 |
-1.21 |
4.56 |
5.151 |
202.393 |
-3.196 |
2.282 |
0.438 |
3.487 |
-3.487 |
5.814 |
-24557.788 |
33 |
-5.76 |
-1.21 |
4.56 |
4.857 |
258.071 |
-3.171 |
2.279 |
0.439 |
3.484 |
-3.484 |
5.177 |
-26122.660 |
34 |
-5.85 |
-1.27 |
4.58 |
5.664 |
212.762 |
-3.635 |
2.291 |
0.436 |
3.561 |
-3.561 |
7.000 |
-26500.452 |
35 |
-5.93 |
-1.32 |
4.61 |
5.958 |
252.609 |
-3.575 |
2.305 |
0.434 |
3.629 |
-3.629 |
7.700 |
-49572.120 |
36 |
-5.23 |
-0.89 |
4.34 |
3.515 |
237.976 |
-3.643 |
2.169 |
0.461 |
3.058 |
-3.058 |
2.848 |
-26501.019 |
37 |
-5.21 |
-0.87 |
4.34 |
3.401 |
233.199 |
-3.637 |
2.169 |
0.461 |
3.042 |
-3.042 |
2.666 |
-26500.962 |
38 |
-5.21 |
-0.86 |
4.35 |
3.229 |
206.205 |
-3.641 |
2.175 |
0.460 |
3.031 |
-3.031 |
2.396 |
-26500.976 |
39 |
-5.22 |
-0.87 |
4.35 |
2.882 |
216.850 |
-3.652 |
2.177 |
0.459 |
3.045 |
-3.045 |
1.908 |
-26501.014 |
40 |
-5.23 |
-0.87 |
4.35 |
2.930 |
290.191 |
-3.895 |
2.177 |
0.459 |
3.051 |
-3.051 |
1.972 |
-27570.289 |
41 |
-5.21 |
-0.86 |
4.35 |
3.125 |
276.250 |
-3.882 |
2.174 |
0.460 |
3.039 |
-3.039 |
2.246 |
-27570.467 |
42 |
-5.29 |
-1.14 |
4.14 |
2.195 |
218.783 |
-3.537 |
2.072 |
0.483 |
3.213 |
-3.213 |
1.162 |
-37938.711 |
43 |
-5.32 |
-1.13 |
4.19 |
4.773 |
249.998 |
-3.526 |
2.093 |
0.478 |
3.224 |
-3.224 |
5.443 |
-37938.674 |
44 |
-5.36 |
-1.39 |
3.97 |
3.386 |
248.611 |
-3.657 |
1.984 |
0.504 |
3.373 |
-3.373 |
2.890 |
-50445.794 |
45 |
-5.27 |
-0.96 |
4.31 |
2.322 |
218.517 |
-3.580 |
2.155 |
0.464 |
3.117 |
-3.117 |
1.251 |
-28132.788 |
46 |
-5.30 |
-1.12 |
4.19 |
2.761 |
196.870 |
-3.594 |
2.093 |
0.478 |
3.210 |
-3.210 |
1.821 |
-28132.866 |
47 |
-5.30 |
-1.01 |
4.29 |
3.959 |
219.714 |
-3.590 |
2.143 |
0.467 |
3.156 |
-3.156 |
3.657 |
-28132.868 |
48 |
-5.30 |
-0.99 |
4.32 |
4.524 |
250.401 |
-3.557 |
2.159 |
0.463 |
3.144 |
-3.144 |
4.740 |
-28132.727 |
49 |
-5.35 |
-1.21 |
4.14 |
3.627 |
249.337 |
-3.706 |
2.072 |
0.483 |
3.279 |
-3.279 |
3.174 |
-40639.867 |
50 |
-5.34 |
-1.08 |
4.26 |
3.406 |
244.373 |
-3.734 |
2.132 |
0.469 |
3.208 |
-3.208 |
2.720 |
-30833.906 |
51 |
-5.28 |
-0.94 |
4.34 |
3.850 |
236.117 |
-3.837 |
2.172 |
0.460 |
3.111 |
-3.111 |
3.412 |
-28547.776 |
52 |
-5.17 |
-0.81 |
4.36 |
4.001 |
206.132 |
-3.715 |
2.180 |
0.459 |
2.992 |
-2.992 |
3.672 |
-27478.836 |
53 |
-5.36 |
-2.60 |
2.76 |
2.491 |
235.239 |
-3.770 |
1.380 |
0.724 |
3.976 |
-3.976 |
2.248 |
-30997.555 |
54 |
-5.22 |
-0.88 |
4.35 |
4.862 |
212.443 |
-3.836 |
2.173 |
0.460 |
3.050 |
-3.050 |
5.440 |
-26937.953 |
55 |
-5.24 |
-0.91 |
4.33 |
3.231 |
195.771 |
-3.407 |
2.165 |
0.462 |
3.070 |
-3.070 |
2.411 |
-25431.563 |
56 |
-5.96 |
-1.43 |
4.53 |
6.110 |
223.819 |
-3.888 |
2.266 |
0.441 |
3.691 |
-3.691 |
8.238 |
-51514.688 |
57 |
-5.31 |
-1.46 |
3.85 |
4.758 |
272.096 |
-4.027 |
1.925 |
0.519 |
3.387 |
-3.387 |
5.879 |
-52584.115 |
58 |
-5.83 |
-1.25 |
4.58 |
5.622 |
266.359 |
-4.107 |
2.288 |
0.437 |
3.541 |
-3.541 |
6.908 |
-28639.355 |
59 |
-5.84 |
-1.19 |
4.66 |
5.019 |
254.722 |
-3.994 |
2.328 |
0.430 |
3.516 |
-3.516 |
5.410 |
-27133.880 |
60 |
-5.96 |
-1.31 |
4.65 |
5.748 |
247.139 |
-3.774 |
2.326 |
0.430 |
3.638 |
-3.638 |
7.102 |
-50009.220 |
61 |
-5.86 |
-1.20 |
4.66 |
4.974 |
197.358 |
-3.521 |
2.328 |
0.430 |
3.532 |
-3.532 |
5.314 |
-24994.981 |
62 |
-5.30 |
-0.93 |
4.37 |
2.503 |
226.866 |
-3.659 |
2.186 |
0.457 |
3.117 |
-3.117 |
1.433 |
-27537.579 |
63 |
-5.46 |
-1.06 |
4.40 |
1.735 |
274.436 |
-4.012 |
2.201 |
0.454 |
3.262 |
-3.262 |
0.684 |
-37337.017 |
64 |
-5.20 |
-1.51 |
3.70 |
5.411 |
246.585 |
-3.642 |
1.849 |
0.541 |
3.355 |
-3.355 |
7.917 |
-36358.769 |
65 |
-5.13 |
-0.79 |
4.34 |
3.235 |
265.187 |
-3.808 |
2.169 |
0.461 |
2.960 |
-2.960 |
2.412 |
-27537.036 |
66 |
-5.23 |
-0.95 |
4.29 |
2.522 |
218.943 |
-3.588 |
2.143 |
0.467 |
3.089 |
-3.089 |
1.484 |
-28547.936 |
67 |
-5.64 |
-1.42 |
4.22 |
1.434 |
224.491 |
-4.022 |
2.108 |
0.474 |
3.527 |
-3.527 |
0.488 |
-29585.673 |
68 |
-5.24 |
-0.86 |
4.38 |
2.898 |
263.211 |
-4.049 |
2.188 |
0.457 |
3.052 |
-3.052 |
1.919 |
-29617.507 |
69 |
-5.18 |
-0.79 |
4.39 |
2.992 |
245.459 |
-3.642 |
2.194 |
0.456 |
2.982 |
-2.982 |
2.040 |
-26501.278 |
70 |
-5.46 |
-1.06 |
4.40 |
1.735 |
252.032 |
-4.012 |
2.201 |
0.454 |
3.262 |
-3.262 |
0.684 |
-37337.017 |
71 |
-5.44 |
-1.04 |
4.40 |
1.959 |
267.979 |
-4.245 |
2.199 |
0.455 |
3.243 |
-3.243 |
0.873 |
-38406.342 |
72 |
-5.09 |
-0.74 |
4.35 |
3.242 |
267.768 |
-4.244 |
2.176 |
0.460 |
2.913 |
-2.913 |
2.415 |
-28639.730 |
73 |
-5.76 |
-1.26 |
4.49 |
2.414 |
260.171 |
-5.440 |
2.247 |
0.445 |
3.509 |
-3.509 |
1.297 |
-42499.229 |
74 |
-5.42 |
-0.93 |
4.49 |
1.971 |
272.458 |
-4.648 |
2.245 |
0.445 |
3.178 |
-3.178 |
0.865 |
-33771.888 |
75 |
-5.57 |
-1.16 |
4.41 |
2.075 |
246.759 |
-4.141 |
2.206 |
0.453 |
3.364 |
-3.364 |
0.976 |
-39383.395 |
76 |
-5.22 |
-1.54 |
3.68 |
6.366 |
257.832 |
-2.846 |
1.842 |
0.543 |
3.380 |
-3.380 |
11.002 |
-33842.015 |
77 |
-5.12 |
-1.08 |
4.04 |
4.167 |
201.459 |
-3.304 |
2.021 |
0.495 |
3.098 |
-3.098 |
4.296 |
-26122.732 |
78 |
-5.58 |
-1.44 |
4.14 |
2.740 |
212.020 |
-2.571 |
2.070 |
0.483 |
3.514 |
-3.514 |
1.814 |
-26026.390 |
EHOMO and ELUMO are associated with electron donating ability and electron accepting ability of a molecule, respectively. Higher EHOMO is essential for molecular reaction with nucleophiles while lower ELUMO reacts easily with electrophiles [26]. EHOMO values for comparative molecules, 60 and 61, of active molecules are found -5.96 and -5.86 eV, and comparative molecules, 24 and 78, of active molecules are found, -5.31 and -5.58 eV, respectively (Table 2). According to these results, the electron donating trends for comparative molecules can be written as: 60>61 for active molecules and 78>24 for inactive molecules. Also, EHOMO values of active molecules are greater than EHOMO values of inactive molecules. EHOMO and ELUMO values of other molecules can be seen in Table 2. There is no significant change in the EHOMO and ELUMO values according to the position of phenyl ring (Figures 6 and 7).
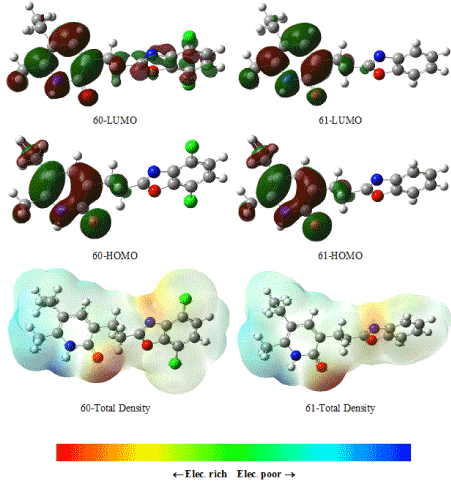
Figure 6. HOMO, LUMO and total density of the inactive molecules for 60 and 61 using DFT/B3LYP/6-311G(d,p) method.
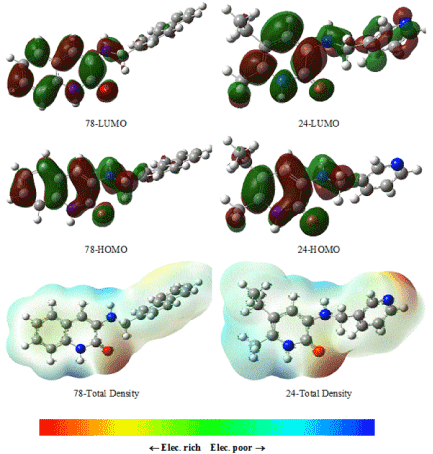
Figure 7. HOMO, LUMO and total density of the active molecules for 78 and 24 using DFT/B3LYP/6-311G(d,p) method.
DE, chemical hardness and softness are closely related to chemical properties of molecules. DE value is smaller when the basis set of atomic orbitals are magnified due to the changing of HOMO, usually to a more negative energy and decreasing in energy of LUMO [27]. More stable molecules have large DE value and less stable molecules have small DE value. DE values of active molecules 60 and 61 that they are more stable were found 4.65 and 4.66 eV, and DE values for inactive molecules 24 and 78 that they are less stable were found 4.32 and 4.14 eV, respectively (Table 2).
2021 Copyright OAT. All rights reserv
The chemical hardness and softness are common used in chemistry for explaining stability of compounds. According to Maximum Hardness Principle [26], chemical hardness is a measure of the stability of chemical species. The chemical hardness is just half the energy gap between the EHOMO and ELUMO (eq. 2). If a molecule has a large energy gap, it is called hard and other wise is called soft [28]. Softness is a measure of the polarizability and soft molecules give more easily electrons to an electron acceptor molecule or surface [23]. The calculated chemical hardness, softness and energy gap are given in Table 2.
The average values of the HOMO and LUMO energies have been defined as the chemical potential (µ). The negative of the chemical potential was known as the electronegativity (c) (eq. 3). The chemical potential, electronegativity and hardness are descriptors for the predictions about chemical properties of molecules [29]. Electronegativity that represents the power to attract the electrons of chemical species is a useful quantity in the prediction of inhibitive performance of molecules [23]. The electronegativity values of active molecules 60 and 61 are more than those of in active molecules 24 and 78 (Table 2).
The total electronic charge (TNC) values were calculated with the 6-311G(d,p) basis set of B3LYP method. TNC values of active molecules are found lower than those for inactive molecules (Table 2).
HOMOs and LUMOs shapes of inactive and active molecules for 2-pyridinone derivatives are also shown in Figure 6 for active template molecules and Figure 7 for inactive template molecules. As seen from the Figures 6 and 7 electron density is concentrated much more in the vicinity of oxygen atoms for active and inactive molecules.
Statistical analysis
The descriptor variables of 78 molecules which are called EHOMO, ELUMO, Energy Gap, Chemical Hardness, Softness, Chemical Potential, Dipole Moment, Total Negative Charge, Molecular Volume, SEZPE, Electro Negativity, Global Electrophilicity and dependent variable called IC50 are investigated in order to determine which descriptors have impact on dependent variable called IC50. It is a fact that the data set has a characteristic of having both linearity and non-linearity among descriptors and between descriptors and dependent variable. Therefore, while Linear Discriminant Analysis (LDA) is conducted in order to determine which attributes have impact on dependent variable for the linear part, Artificial Neural Networks (ANNs) is run for the non-linear part in order to determine which attributes have impact on it.
In order to run LDA, the dependent variable is split into two non-overlapping data sets. While the first group consists of 58 molecules whose values range between 0.019 and 2.850, the second group composes of 20 molecules whose values are relatively high values and alter between 3.7 and 300.00. Therefore, LDA calculated function which separates the first group from the second group is given in standardized coefficient form as follows:
0.958 * Molecular Volume — 0.252 * Global Electrophilicty + 0.152 * ELUMO
The 39 percent of the total variance is explained by the linear discriminant function given above and its level of significance measured by Wilk’s lamba is 0008<0.05 shows that the model is statistically significant. On the other hand, linear discriminant function calculated above has a low misclassification rate of 23 percent. Also, when cross-validated, its misclassification rate hits 25 percent, which means that when one of the data used for constructing modeling repeatedly is excluded, the correct classification rate of the constructed model for the excluded observation hits 75 percent. In other words, when a new molecule arrives in data set, it is possibly being classified with 75 percent accuracy. All computations are done using SPSS 20.0 version [30].
For the non-linear part, Artificial Neural Networks (ANNs) with multilayer perceptron using sigmoid activation function is conducted using SPSS 20.0 version. The descriptors except the ones used in LDA, namely, EHOMO, Energy Gap, Softness, Hardness, Chemical Potential, Dipole Moment, Total Negative Charge, SEZPE, Electro Negativity are employed in order to predict which descriptors have impact on dependent variable IC50. The data set is split into two non-overlapping sets. The first of which is called training set and the second one is called the test set whose partitions are 70 percent and 30 percent, respectively. Dipole Moment, Hardness, EHOMO and Electro Negativity are determined as the most important descriptors with 0.64 coefficient of determination.
As a result, due to the complex structure of the data set, it requires the investigation to be conducted including both linearity and non-linearity analysis which are Linear Discriminant Analysis (LDA) and Artificial Neural Networks (ANNs), respectively. While Molecular Volume, Global Electrophilicity and ELUMO are the most significant descriptors and have a power of correctly separating molecules with 77 percent for linear part, Dipole Moment, Hardness, Homo and Electro Negativity are descriptors having impact on dependent variable called IC50 for non-linear part.
In this study, we find the relationship between HIV-1 activity and chemical structure for 2-pyridinone derivatives by using ETM, and also we calculated and discussed quantum chemical parameters of 2-pyridinone derivatives such as the energy of the highest occupied molecular orbital, the energy of the lowest unoccupied molecular orbital, HOMO–LUMO energy gap, chemical hardness, softness, electronegativity, chemical potential, dipole moment, global electrophilicity, sum of the total negative charge (TNC) and sum of electronic and zero-point energies (SEZPE) quantum-mechanical calculations by using B3LYP method with basis set of the 6-311G(d,p) in order to find molecular properties.
Based on those calculations, two statistical models conducted for the determination of statistically significant descriptors for the linear and non-linear parts of the data set are Linear Discriminant Analysis (LDA) and Artificial Neural Networks (ANNs), respectively. Due to the complex structure of data set, linearly correlated and non-linearly correlated descriptors are separately used in order to find which attributes having impact on dependent variable. While Molecular Volume, ELUMO and Electrophilicity are significant descriptors for linear part determined by LDA, Dipole Moment, Hardness, EHOMO and Electro Negativity are significant descriptors for non-linear part of the data set.
- de Clercq E (1996) Non-nucleoside reverse transcriptase inhibitors (NNRTIs) for the treatment of human immunodeficiency virus type 1 (HIV-1) infections: strategies to overcome drug resistance development. Med Res Rev 16: 125-157. [Crossref]
- Debnath AS, Radigan L, Jiang S, Lindsley F (1999) Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J Med Chem 42: 3203-3209.
- De Clercq E (2000) Novel compounds in preclinical/early clinical development for the treatment of HIV infections. Rev Med Virol 10: 255-277.
- Temin HM, Mizutani S (1970) RNA-dependent DNA polymerase in virion of rous sarcoma virus. Nature 226: 1211-1213.
- Nouvel P (1994) The mammalian genome shaping activity of reverse transcriptase. Genetica 93: 191-201. [Crossref]
- De Clercq E (1993) HIV-1 specific RT inhibitors: Highly selective inhibitors of human immunodeficiency virus type 1 that are specifically targeted at the viral reverse transcriptase. Med Res Rev 13: 229-258.
- Dimoglo AS, Chumakov YM, Dobrova BN, Saracoglu M (1997). Electron-topological of the Structure-antitumor activity relationship of thiosemicarbazone derivatives. Arzn-Forsch-Drug Res 47: 415-419.
- Kandemirli F, Saracoglu M, Kovalishyn V (2005). Human acetylcholinesterase inhibitors: Electronic-topological and neural network approaches to the structure-activity relationships study. Mini-Rev Med Chem 5: 479-487.
- Dimoglo AS, Gorbachov MY, Lesnik TI, Saracoglu M, Güzel Y, et al. (1997) Investigation of the relationship between chemical structure and anti-HIV-1 activity in a class of nucleoside analogues: Electron topological approach. Curr Med Chem 4: 23-34.
- Saracoglu M, Kandemirli SG, Basaran A, Sayiner H, Kandemirli F (2011) Investigation of structure-activity relationship between chemical structure and CCR5 antiHIV-1 activity in a class of 1-[N-(methyl)-N-(phenylsulfonyl) amino]-2-(phenyl)-4-[4-(substituted) piperidin-1-yl] butanes derivatives: The electronic-topological approach. Curr HIV Res 9: 300-312.
- Kandemirli F, Kovalishyn V, Kandemirli SG (2007) Electron-topological method of the non-nucleoside HIV-1 RT inhibitors study: Structure-activity relationships. Curr HIV Res 5: 449-458.
- Garg R, Gupta SP, Gao H, Babu MS, Debnath AS, et al. (1999) Comparative quantitative structure-activity relationship studies on anti-HIV drugs. Chem Rev 99: 3525-3601.
- Saari WS, Wai JS, Fisher TE, Thomas CM, Hoffman JM, et al. (1992) Synthesis and evaluation of 2-pyridinone derivatives as HIV-1-specific reverse transcriptase inhibitors. 2. Analogues of 3-aAminopyridin-2(1H)-one. J Med Chem 35: 3792-3802.
- Saari WS, Hoffman JM, Wai JS, Fisher TE, Rooney CS, et al. (1991) 2-Pyridinone derivatives: A new class of nonnucleoside, HIV-1- specific reverse transcriptase inhibitors. J Med Chem 34: 2922-2925.
- Dimoglo AS (1985) Compositional approach to electronic structure description of chemical compounds, oriented on computer analysis of structure-activity relationships. Khim-Pharm Zhurn 4: 438-444.
- Shvets NM (1993) Applied program system for the prognosys of biological activity of chemical compounds: Development and use. Comp Sci J Moldova 1: 101-110.
- Shvets NM (1997) The study of data and control flows and the user interface organization in an applied system used in chemistry and medicine for the biological activities prediction. Comp Sci J Moldova 3: 301-311.
- Dimoglo AS, Vlad PF, Shvets NM, Coltsa MN (2001) Structure-ambergris odour relationships investigation in a mixed series of decalin and non-decalin compounds the electronic-topological approach. New J Chem 25: 283-288.
- Dimoglo AS, Shvets NM, Tetko IV, Livingstone DJ (2001) Electronic-topological investigation of the structure-acetylcholinesterase inhibitor activity relationship in the series of n-benzylpiperidine derivatives. QSAR 20: 31-45.
- Kandemirli F, Tokay N, Shvets NM, Dimoglo AS (2003) Electronic-Topological Study of the Structure-Activity Relationships in a Series of Steroids with Mineralocorticoid Binding Affinity. Arzneimittelforschung 53: 133-138.
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. (2004) Gaussian Inc, Wallingford, CT.
- Parr RG, Pearson RG (1983) Absolute hardness: companion parameter to absolute electronegativity. J Am Chem Soc 105: 7512-7516.
- Kaya S, Kaya C, Guo L, Kandemirli F, Tüzün B, et al. (2016) Quantum chemical and molecular dynamics simulation studies on inhibition performances of some thiazole and thiadiazole derivatives against corrosion of iron. J Mol Liquids 219: 497-504.
- Chattaraj PK, Sarkar U, Roy DR (2006) Electrophilicity index. Chem Rev 106: 2065-2091. [Crossref]
- Ebenso EE, Kabanda MM, Arslan T, Saracoglu M, Kandemirli F, et al. (2012) Quantum chemical investigations on quinoline derivatives as effective corrosion inhibitors for mild steel in acidic medium. Int J Electrochem Sci 7: 5643-5676.
- Rank A (2001) Orbital Interaction Theory of Organic Chemistry. (2nd Edn), Wiley & Sons: New York.
- Pearson R (1993) The principle of maximum hardness. Acc Chem Res 26: 250-255.
- Pearson RG (1986) Absolute electronegativity and hardness correlated with molecular orbital theory. Proc Natl Acad Sci U S A 83: 8440-8441. [Crossref]
- Özalp A, Kökbudak Z, Saracoglu M, Kandemirli F, Ilhan IÖ, et al. (2015) Synthesis and theoretical study of the novel 2-oxopyrimidin-1(2H)-yl-amides derivatives. Chem Sci Rev Lett 4: 719-728.
- IBM (2016) SPSS 200 Manual, USA.











