Objective: To characterize group B streptococcus (GBS) isolated from patients at the Kosar Maternity Hospital in Ghazvin, a labor-dominated industrial city of Iran for their serotypes, virulence factors, and antibiotic resistance.
Materials and Methods: A total of 270 samples were collected from vaginal swabs, midstream urine, and rectal swabs of 90 adult participants throughout 2019-2021 during 35-37 weeks of gestation. Standard bacteriological methods, CAMP test, and amplification of the CFB gene encoding CAMP factor by polymerase chain reaction using specific primers were carried out to identify the GBS isolates. The overall prevalence and identification of GBS serotypes and virulence factors were studied using multiplex PCR. The isolates were subjected to antimicrobial susceptibility tests.
Results: The preliminary identification tests revealed a 17.6% prevalence of GBS among the patients. The multiplex PCR led to the identification of the serotypes Ia (25%), Ib (50%), and II (25%). Three distinct virulence gene profiles of cylE, lmb, and bca were identified. The serotype (Ia) with three virulence factors of CylE, lmb, bca isolated from rectal samples was resistant to all antibiotics except vancomycin. The serotype Ib isolated from urine samples was resistant to all antibiotics except Ampicillin, while serotype II was sensitive to all the antibiotics except cefotaxime.
Conclusions: The prevalence of colonization and antibiotic resistance of GBS is relatively high in mothers who participated in the present study. Resistance to commonly administered antibiotics indicates that other measures such as vaccination are potentially useful to prevent and control GBS in this population. Pregnant mothers should be screened for GBS colonization and antimicrobial susceptibility testing. This study has provided initial data on the genotypes and prevalence of virulence genes and antibiogram profile in GBS isolates obtained at the Kosar Maternity Hospital, Ghazvin, Iran.
streptococcus agalactiae, group b streptococcus, prevalence, serotype, genotypes, virulence, antibiotic resistance
S. agalactiae, Group B Streptococcus (GBS), is part of the human intestinal microbiota, and one of the major causative agents of severe infections in pregnant women. The colonization of GBS is generally asymptomatic in the form of a member of the gastrointestinal or vaginal microbiota but can cause life-threatening infections [1]. GBS is also known to cause morbidity and mortality in high-risk populations, neonates, and nonpregnant adults particularly in immunocompromised and Diabetics [2,3]. S. agalactiae colonizes the genital tract from the intestine, and is transmitted to the newborn. 15-35% of pregnant women are vaginally colonized, and about 1-2% of the colonized neonates develop severe infections [4]. Approximately 25% of pregnant women harbor rectal or vaginal S. agalactiae [5] which can be vertically transmitted to the fetus leading to early onset of sepsis, meningitis, and pneumonia [4]. Urinary tract infections are another manifestation of GBS disease that may be associated with a variety of GBS serotypes [6]. GBS bacteriuria in women during pregnancy is a risk factor for late gestational maternal colonization with GBS and early-onset neonatal GBS infection [7]. GBS vaginal colonization rates in many individual countries are unknown. Estimates in countries of GBS vaginal colonization can be highly variable. In pregnant women, GBS can be transmitted to newborns causing severe infections. Capsular serotype and virulence factors are the major determinants of neonatal disease severity. These factors are necessary for host-bacterial cell interaction [8]. Virulence factors facilitate the disease-causing ability of GBS. Capsular polysaccharides are the most important virulence factors that vaccine development strategies target. A range of virulence factors has been attributed to GBS that facilitates its ability to cause disease. Some of these virulence factors include capsular polysaccharides, regulatory proteins, surface-localized proteins, and toxins [9]. Two types of carbohydrate antigens were defined in the GBS wall. The group B antigen, common to all strains, and the specific capsular polysaccharide antigen that allows its classification into 10 serotypes (Ia, Ib, II-IX), with different geographic distribution [3]. The severity of the neonatal disease is determined by the capsular serotype and virulence factors such as the polysaccharide capsule, encoded by the cps gene, protein C, which includes the α-subunit of C protein (bca gene), and β-subunits of the C protein (bac), resistance to protease immunity protein (rib); the laminin binding protein Lmb (lmb gene), secreted fibrinogen-binding protein FbsA (fbsA gene), fibrinogen-binding protein FbsB (fbsB gene), the cyl operon encoding a β-hemolysin hyaluronidase (hylB gene), the CAMP factor (cfb gene) that promote pathogen entry into host cells and facilitate its intracellular survival and spread, and the C5a peptidase (scpB gene) [10,11]. Penicillin is recommended as the first choice to prevent GBS infection in newborns, for intrapartum prophylaxis in pregnant women. Drugs like macrolides are the therapeutic alternative for penicillin-intolerant women. Increasing loss of susceptibility to these antibiotics among GBS isolates has become a global concern [12]. A reduced number of clinical isolates with penicillin susceptibility with a tendency to multidrug resistance, has been reported between 2005–2006 and 2012–2013 in Japan [13]. Test for GBS is recommended by the Center for Disease Prevention and Control (CDC) in all women between 35 and 37 weeks of gestation by vaginal (anterior third) and anorectal swabbing [14]. This work aimed to determine the degree of GBS colonization in pregnant women, the serotypes distribution, antimicrobial resistance pattern, and virulence-associated genes in a labor-dominated industrial city from the patients attending a government maternity hospital.
Study design
This study was conducted from October 2019 to March 2021 at Kosar maternity hospital of Ghazvin, Iran. The study was approved by the Ethics Committee of Ghazvin University of Medical Sciences. All participants completed written informed consent. In this study, 270 samples were collected from 90 patients. Rectal, vaginal, and urine samples were collected from each patient at 35–37 weeks of pregnancy as recommended by the Center for Disease Control [14].
GBS isolation from urine samples
A clean-catch midstream urine specimen for conventional quantitative culture was collected from each patient during the planned visit to the midwife outpatient clinic. Urine samples were seeded on a 5% blood agar plate and read after incubation at 35 oC for 24 or 48 h depending on the initial growth of bacteria.
Sampling and culture methods
The samples were taken from the anterior third of the vagina with a swab and simultaneously with another swab from the anorectal area. The patients did not receive antimicrobials days before and during the sampling. The vagino-rectal swab was seeded in 1–2 mL of Todd-Hewitt broth supplemented with colistin (10 μg mL− 1) and nalidixic acid (15 μg mL− 1). The sown stock was grown 18–24 h in an incubator under microaerophilic conditions (5% CO2) at 35 °C [10]. After incubation, it was transferred to a 5% sheep blood agar plate with a seeding technique for isolation. The plates were incubated in a micro-aerobic atmosphere for 24 h. Suspicious β or γ hemolytic colonies were taken from the plates. Identification of GBS was performed based on colony morphology, β-hemolytic reaction, Gram-stain, hippurate hydrolysis, and cAMP factor using the standard test of CAMP (Christie, Atkins, and Munch Petersen test).
Molecular identification of GBS
500 μL of Todd–Hewitt broth containing each of the vaginal, rectal, or urine samples were taken in 1.5 mL microtubes. DNA was extracted using GeneAll DNA extraction kit (Seoul, Korea) according to the manufacturer's recommendations. To identify GBS, amplification of the 153bp cfb gene (encoding the CAMP factor that is conserved in all GBS isolates) was performed using the specific primers F: 5’TTTCACCAGCTGTATTAGAATA-3’ and R: 5’ GTTCCCTGAACATTATCTTTGAT-3’. Alternatively, scpB coding for C5a peptidase was amplified to produce a 255bp band confirming GBS [15].
Serotyping and determination of virulence genes
Multiplex PCR was used to screen for the carriage of serotype and virulence genes including α- (bca) and β-subunits of the C protein (bac), resistance to protease immunity protein (rib), surface immunogenic protein (sip), b-hemolysin/cytolysin (cylE), laminin-binding protein (lmb), a surface protein of group B streptococcus (spb), C5a peptidase (scpB) and hyaluronidase (hylB). Molecular typing for S. agalactiae was determined by thr detection of cps clusters using specific primers [16] (Table 1) with some modifications described by Kannika et al. (2017) [17]. The isolates were screened for the virulence factors in multiplex PCR assays using primers (Table 2) and conditions published previously [15,18-22].
Table 1: Size (in base pairs, bp) and type of the PCR amplification products expected for each serotype (Ia to IX)
No. |
GBS capsular gene |
Sequence 5′ → 3′ |
Serotype |
Size |
1 |
cpsL |
F: CAATCCTAAGTATTTTCGGTTCATT
R: TAGGAACATGTTCATTAACATAGC |
Ia, Ib,
II-IX
|
688
|
2 |
cpsG |
F: ACATGAACAGCAGTTCAACCGT
R: ATGCTCTCCAAACTGTTCTTGT
CpsG-2-3-6-R: TCCATCTACATCTTCAATCCAAGC |
Ia
II
III
IV
V
VII
IX |
272
272
352
272
272
272
272 |
3 |
cpsI-Ia-6-7-F |
F: GAATTGATAACTTTTGTGGATTGCGATGA |
|
|
4 |
cpsI-6-R |
R: CAATTCTGTCGGACTATCCTGATG |
VI |
470 |
5 |
cpsI-7-9-F |
F: CTGTAATTGGAGGAATGTGGATCG |
|
|
6 |
cpsI-7-R |
R: TGTCGCTTCCACACTGAGTGTTGA |
VII |
179 |
7 |
cpsI-9-R |
R: AATCATCTTCATAATTTATCTCCCATT |
IX |
229 |
8 |
CpsN-5 |
F: ATGCAACCAAGTGATTATCATGTA
R: CTCTTCACTCTTTAGTGTAGGTAT |
V |
582 |
9 |
CpsJ-8 |
F: TATTTGGGAGGTAATCAAGAGACA
R: GTTTGGAGCATTCAAGATAACTCT |
VIII |
438 |
10 |
cpsJ-2-4-F |
F: CATTTATTGATTCAGACGATTACATTGA |
|
|
11 |
cpsJ-2-R |
R: CTCTTTCTCTAAAATATTCCAACC |
II |
465 |
12 |
cpsJ-4-R |
R: CCTCAGGATATTTACGAATTCTGTA |
IV |
538 |
13 |
cpsJ-Ib |
F: GCAATTCTTAACAGAATATTCAGTTG
R: GCGTTTCTTTATCACATACTCTTG |
Ib |
621 |
Table 2: Primers used to amplify the virulence factors
Gene |
Primers |
Amplicon size (bp) |
bca |
Fwd: 5'-TAACAGTTATGATACTTCACAGAC-3'
Rvs: 5-'ACGACTTTCTTCCGTCCACTTAGG-3' |
535 |
bac |
Fwd:5'-CTATTTTTGATATTGACAATGCAA-3'
Rvs: 5'-GTCGTTACTTCCTTGAGATGTAAC-3' |
592 |
bca |
Fwd: CAGGAGGGGAAACAACAGTAC
Rvs: GTATCCTTTGATCCATCTGGATACG |
183 |
bac |
Fwd: TGTAAAGGACGATAGTGTGAAGAC
Rvs: CATTTGTGATTCCCTTTTGC |
530 |
rib |
Fwd:5'-CAGGAAGTGCTGTTACGTTAAAC-3'
Rvs:5'-CGTCCCATTTAGGGTCTTCC-3' |
369 |
sip |
Fwd: ATCCTGAGACAACACTGACA (position 263–282)
Rvs: TTGCTGGTGTTTCTATTTTCA (position 340–320) |
78 |
cylE |
Fwd: TGA CAT TTA CAA GTG ACG AAG (position 1541–1561)
Rvs: TTG CCA GGA GGA GAA TAG GA (position 1808–1789) |
268 |
cylE |
Fwd:TCGGAACAAGTAAAGAGGGTTCGG
Rvs: GGGTTTCCACAGTTGCTTGAATGT
(GenBank accession numbers AF09378.2 and AF157015.2) |
130 |
lmb |
Fwd: GACGCAACACACGGCAT
Rvs: TGATAGAGCACTTCCAAATTTG |
300 |
scpB |
Fwd: ACAATGGAAGGCTCTACTGTTC
Rvs: ACCTGGTGTTTGACCTGAACTA |
255 |
hylB |
Fwd:5′-CATACCTTAACAAAGATATATAACCCAAA-3′
Rvs:5′-AGATTTTTTAGAGAATGAGAAGTTTTTT-3′ |
950 |
hylB |
Fwd:5′- TGTCTCCGAGGTGACACTTGAACT -3′
Rvs:5′- TTGTGTTGTGACGGGTTGTGGATG-3′
(GenBank accession numbers Y15903 and U15050) |
124 |
Antimicrobial susceptibility tests
Antibiotic susceptibility testing was performed by the disc diffusion (Kirby-Bauer) method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [23]. Streptococcus pneumonia (ATCC 49619) was used as a control. The antibiotic discs procured from Himedia (India) were: Penicillins [(penicillin G (P) 10 units and ampicillin (AMP), 10 µg]; cephalosporins [ceftriaxone (CRO), 30 µg; cefotaxime (CTX), 30 µg; cefepime (FEP), 30 µg;], a macrolide [erythromycin (E), 15 µg], a glycopeptide [vancomycin (VA), 30µg], a lincosamide [clindamycin (DA), 2 µg], a phenicole (chloramphenicol) and tetracycline (TC), 30 µg.
Statistical analysis
The SPSS statistical analysis package (version 21.0 for Windows) was used to analyze the significance of the results. Chisquare tests of independence were performed to assess possible associations between the variables. The significance level was set at p <0.05.
Isolation and identification of GBS
Bacteriological tests followed by CAMP test and amplification of the gene encoding CAMP factor called cfb, by polymerase chain reaction using specific primers led to the identification of a 17.6% prevalence of GBS among the patients.
Serotyping and determination of virulence genes
Overall, three serotypes viz., Ia (Figure 1), Ib (Figure 2), and II (Figure 3) were identified. Serotypes Ia, Ib, and II accounted for 25%, 50%, and 25% (Figure 4) of the total isolates. Three distinct virulence gene profiles were identified (Figure 5). cylE was common in all the isolates, and lmb was predominant in 75% while bca accounted for 87.5% (Figure 6). Serotype II was detected from vaginal samples with virulence factors CylE and lmb. Serotype Ia was from rectal samples and serotype Ib with virulence factors CylE, lmb, and bca were from both vaginal and urine samples (Figure 5).
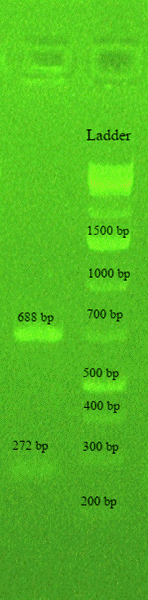
Figure 1: Gel electrophoresis of the multiplex PCR amplification products showing two bands of 688 (cpsL) and 272 (cpsG) bp confirming serotype Ia of a rectal isolate of GBS
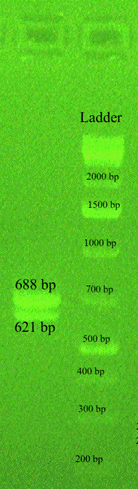
Figure 2: Gel electrophoresis of the multiplex PCR amplification products showing two bands of 688 (cpsL) and 621 (cpsJ) bp confirming serotype Ib of a urinal isolate of GBS

Figure 3: Gel electrophoresis of the multiplex PCR amplification products showing two bands of 688 (cpsL) and 465 (cpsJ) bp confirming serotype II of a vaginal isolate of GBS
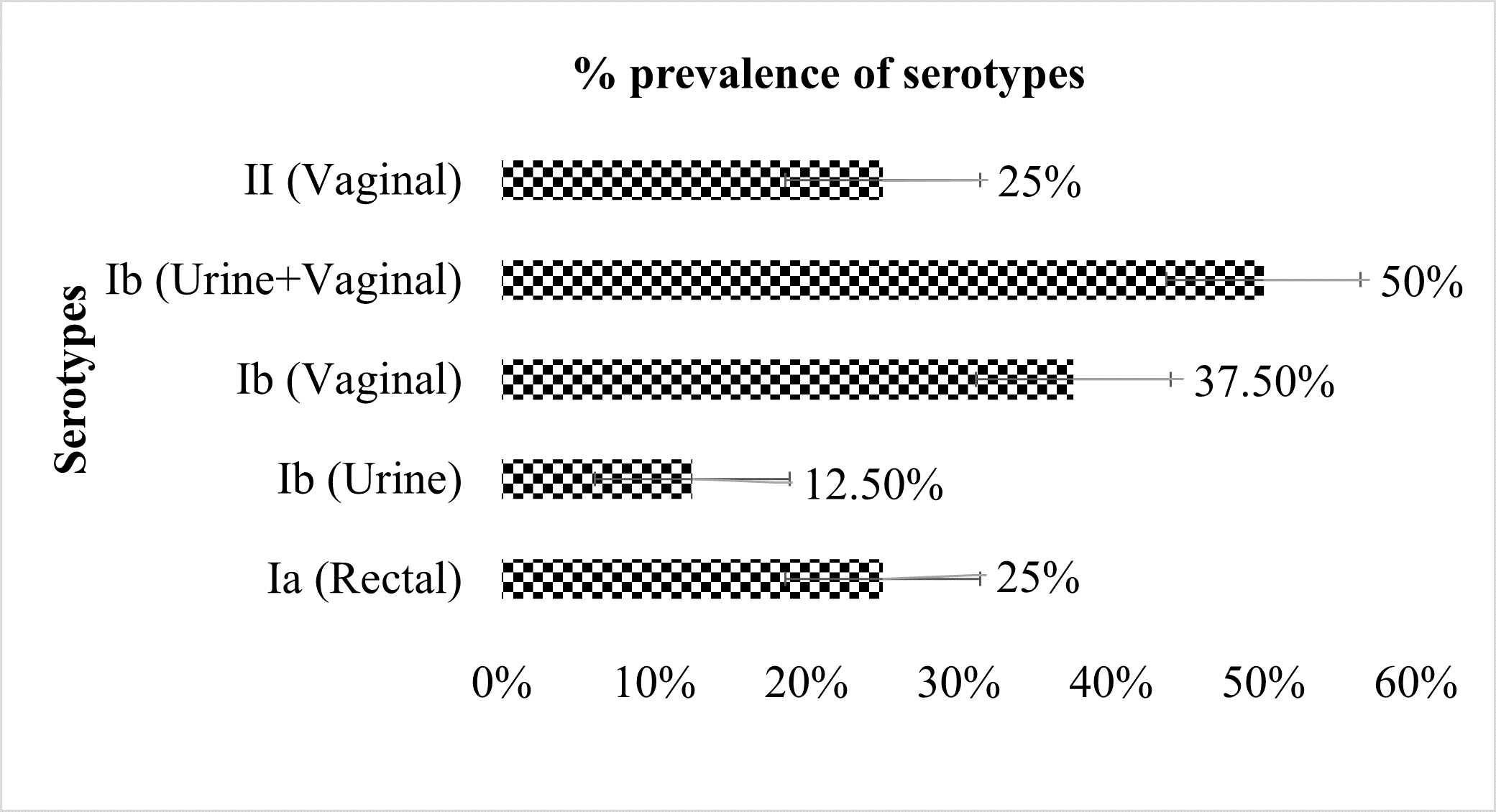
Figure 4: Distribution of GBS serotypes isolated from vagina, rectum, and urine samples
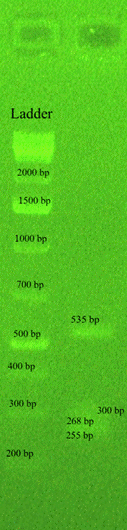
Figure 5: Gel electrophoresis of the multiplex PCR amplification products from a GBS serotype Ib of a urinal isolate showing four bands of 535, 300, 268, and 255 base pairs pertaining to the virulence factors bca, lmb, cylE, and scpB respectively
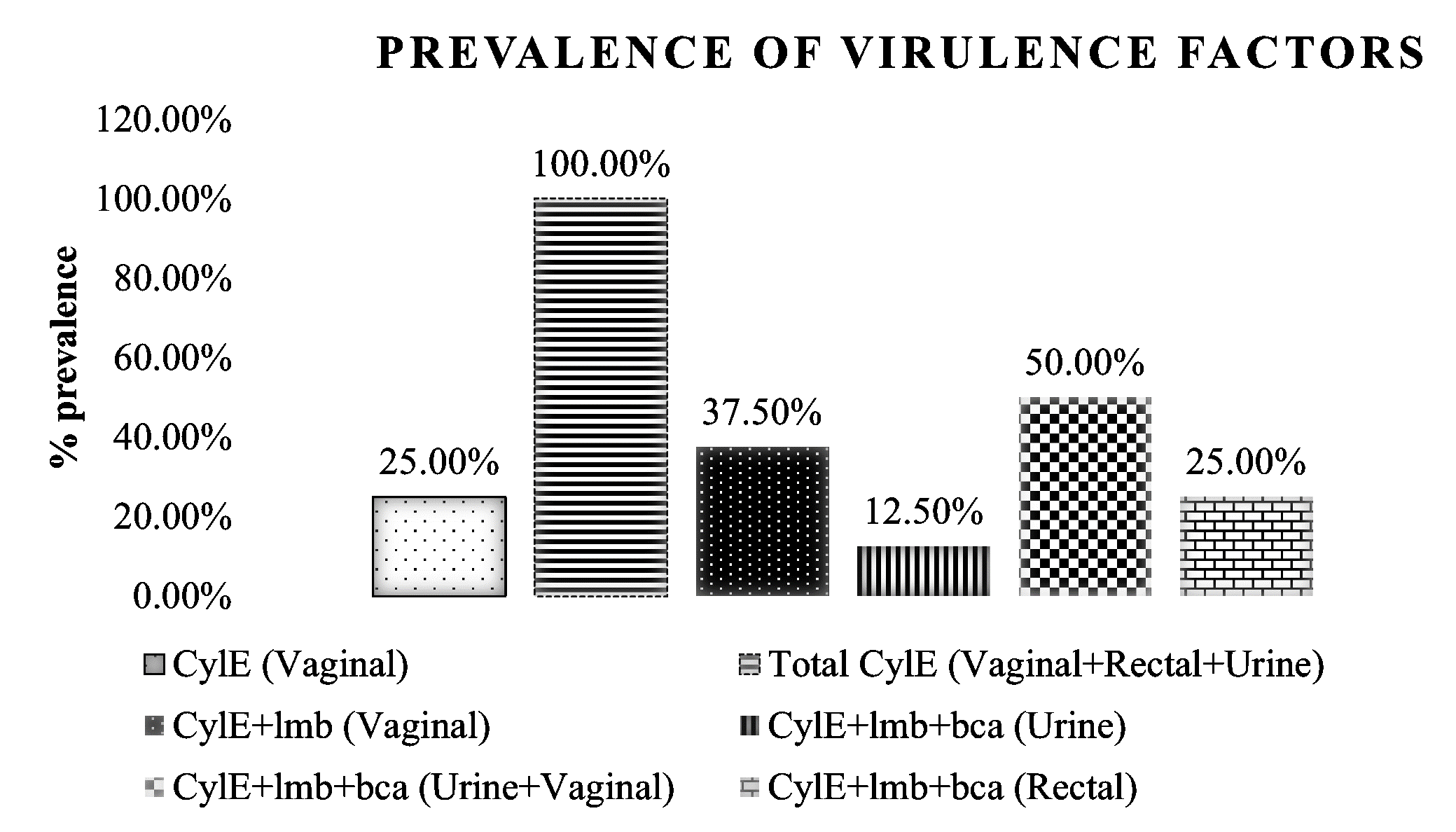
Figure 6: Distribution of virulence factors in GBS isolates from different sampling sources
Antimicrobial susceptibility tests
The serotype Ib from urine samples was resistant to all antibiotics except Ampicillin. This serotype accounted for 2.2% of the total samples or 12.5% of the positive samples. Serotype (Ia) with three virulence factors CylE, lmb, bca from rectal samples was 4.4% of the total patients or equivalent to 25% of positive samples. These samples were resistant to all antibiotics except vancomycin. Also, serotype (Ib) with two virulence factors CylE and lmb were isolated from vaginal samples in 6.6% of ninety patients or 37.5% of positive samples. All the isolates were resistant to the range of the antibiotics tested except for Ceftriaxone in 18.75% of the positive samples. Overall isolates were resistant to penicillin (75%), Ampicillin (43.75%), Chloramphenicol (75%), Clindamycin (75%), Cefepime (75%), Ceftriaxone (56.25%), Erythromycin (75%), Vancomycin (50%), Tetracycline (75%), and Cefotaxime (100%) (Table 3).
Table 3: Antimicrobial susceptibility of group B Streptococcus isolates
| Antimicrobials
|
Susceptibility
|
| |
S (%)
|
I (%)
|
R (%)
|
Penicillin |
25 |
0 |
75 |
Ampicillin |
56.25 |
0 |
43.75 |
Chloramphenicol |
25 |
0 |
75 |
Clindamycin |
25 |
0 |
75 |
Cefepime |
25 |
0 |
75 |
Ceftriaxone |
43.75 |
0 |
56.25 |
Erythromycin |
25 |
0 |
75 |
Vancomycin |
50 |
0 |
50 |
Tetracycline |
25 |
0 |
75 |
Cefotaxime |
0 |
0 |
100 |
S: Susceptible; I: Intermediate; R: Resistant
Streptococcus agalactiae is a contagious pathogen that causes bovine mastitis worldwide, resulting in considerable economic losses. Studies have shown that vaginal infection with GBS during pregnancy increases the risk of vertical transmission of the infection to infants and the likelihood of late infection [4]. Late-onset infection generally presents as meningitis, sepsis, and pneumonia in infants, and in 90% of cases shows symptoms within the first 24 hours. Based on a systematic review and meta-analysis of the results of the studies, the prevalence of GBS colonization in Iranian pregnant women is estimated to be 13.65% [24]. The prevalence in other developing countries such as India, Turkey, Thailand, Kuwait, Saudi Arabia, and Nigeria is 2.3, 9.2, 12.9, 16.4, 13.9, and 19.5 percent, respectively [25]. Differences in laboratory standards and methods of diagnosis, health and medical policies of countries, preventive measures associated with ethnic differences, geographical area, differences in sampling locations, bacteriological detection methods, statistical differences in the study population, the skills of laboratory staff, and sexual partners can be the cause of differences in other countries. The prevalence of GBS colonization is higher in developed countries, such as Italy (16.5), Brazil (14.6), Canada (11.6), the United Kingdom (10.5), Greece (6.6%), and Israel (6.5%) [26-31]. Based on the results of the meta-analysis, the sampling area and prevalence of GBS colonization in Iranian pregnant women were statistically significant (p = 0.001). The prevalence of vaginal, vaginorectal, and anovaginal colonization of GBS were 11.96%, 13.62%, and 25.63% respectively [24]. In Iran, the pattern of different antibiotic resistance and differences in antigen, and pathogenicity of different serotypes in different geographical areas can be attributed to several factors [26-34]. Also, in the evaluation conducted by Fazeli et al. [35], the prevalence of GBS colonization in rural pregnant women was high. More contact with animals, livestock, and its products can be effective factors in this regard [36]. Antibiotic susceptibility of bacteria reported by the disc diffusion method indicated that 100% of the cases were resistant to penicillin and ampicillin while 26.9% and 42.1% were resistant to erythromycin and clindamycin [37]. In a study in the United States, all isolates were sensitive to penicillin, ampicillin, and cefazolin, and resistance rates of 25.6% and 12.7% were reported for erythromycin and clindamycin, respectively [38]. In the present study, 270 samples were taken from 90 pregnant patients aged 37-35 weeks volunteered with a mean age of 30 years. The urinary isolates, which accounted for 12.5% of the total isolates, were resistant to all antibiotics except ampicillin. However, the range of antibiotic resistance of other isolates was very wide and they were sensitive to a fewer number of antibiotics, which is worth considering strict controls on the overuse of over-the-counter supply of antibiotics. Earlier the sensitivity of GBS in Iranian pregnant women was reported as 97.2% to ampicillin, 80.5% to erythromycin, 83.4% to clindamycin [39], and cefazolin 76.5% [24]. Therefore, according to studies on antibiotic resistance and susceptibility to GBS, penicillin has been selected for the prevention and treatment of GBS infection, which is usually prescribed in early labor. Erythromycin and clindamycin are prescribed to pregnant women at risk for anaphylaxis instead of penicillin or cefazolin [40]. While penicillin is a good choice for women who are not at risk for anaphylaxis [41], nitrofurantoin can also be effective in the symptomatic and asymptomatic treatment of S. agalactiae. Vancomycin can be used if the drug is resistant to these antibiotics [35]. Following the recommendations of the CDC and the use of prophylactic antibiotics in leading countries has seen a 70% reduction in the prevalence of infection in infants [27]. This has been confirmed in Iranian studies [26,31,42,43]. The incidence of early-onset neonatal disease has been reduced by intrapartum antibiotic prophylaxis. Despite reduced penicillin susceptibility among GBS isolates, penicillin G is still the mainstay of the therapy. Some isolates exhibited resistance to non-beta-lactam antibiotics, including clindamycin, erythromycin, and fluoroquinolones [3]. The susceptibility to penicillin and trimethoprim/sulfamethoxazole of S. agalactiae was reported while the strains isolated from patients aged 45–54 were resistant to clindamycin and erythromycin [44]. Penicillin and ceftriaxone have been reported to exert cidal effects on GBS isolates suggesting penicillin as the first choice for antibiotic prophylaxis and treatment of GBS infections [12]. Our study showed a decreased level of GBS sensitivity to antibiotics as compared to the previous reports indicating a demand for alternative measures. This indicates that in order to prevent the invasion of the disease, and reduce the GBS burden, it is essential to develop and implement some strategies to identify and treat hosts with narrow spectra antimicrobials or vaccine.
Surface proteins mostly act as adhesins, which may also play a role in escaping the immune system. These include peptidase C5a (ScpB), laminin-binding protein (Lmb), α (Bca) and β (Bac) subunits of protein C. In this study, cylE virulence factor was observed in all isolates. This hemolytic factor is a major determinant of colonization and pathogenesis in group B streptococci. GBS hemolytic pigment is also known as Granadaene. There are many ambiguities in our understanding of residual granadaene, including its biosynthetic profile in bacterial cells and the reasons why GBS may have evolved to produce this potent toxin. Such insights can improve our understanding of this important GBS virulence factor as well as other similar and potentially toxic microbial lipids. cyl operon (cylX-K) is essential for the production of hemolytic pigments in GBS [45] and is an important factor in enhancing the virulence of beta-hemolysin/cytolysin, which is cytotoxic to host cells. The expression of operon seal in GBS is sufficient to induce the production of Granadaene pigment in Lactococcus lactis, a non-hemolytic gram-positive bacterium. The purified pigment from L. lactis is structurally and cytolytically identical to the granadaene extracted from GBS, indicating that the operon seal is sufficient to produce granadaene in a heterogeneous host [45]. These findings provide a greater understanding of the biosynthesis and evolutionary basis of a key GBS virulence factor and suggest that such potentially toxic lipids may be encoded by other bacteria. Analyzes show that cyl operon genes are present in a diverse range of gram-positive bacteria, and it can be argued that pigment biosynthesis in living and free bacteria probably evolved as a defense mechanism against competing bacteria. The present study revealed a prevalence of virulence genes among the GBS isolates with most of the isolates carrying at least three virulence genes that code for surface-localized proteins, regulatory proteins, and toxins. Serotype (II) was detected from vaginal samples with a virulence factor CylE that was sensitive to all antibiotics except Cefotaxime. This serotype accounted for 4.4% of the total samples or 25% of the positive samples. Serotype (Ib) with three virulence factors CylE, lmb, bca from urine samples was 2.2% of the total patients or equivalent to 12.5% of positive samples. These samples were resistant to all antibiotics except Ampicillin. Also, serotype (Ib) with two virulence factors CylE and lmb were isolated from vaginal samples in 6.6% of ninety patients or 37.5% of positive samples. Half of these samples were resistant to all antibiotics and the other half were only sensitive to Ampicillin and Ceftriaxone and resistant to other antibiotics. Serotype (Ia) with three virulence factors CylE, lmb, bca of rectal samples accounted for 4.4% or 25% of positive samples. These samples were resistant to all antibiotics except Vancomycin.
The serotypes are distributed regionally in the world. Ia, Ib, II, III, and V are the most common serotypes accounting for more than 85% of serotypes in the world [46]. The invasive disease caused by GBS manifests in a wide spectrum of clinical findings. Serotypes Ia, Ib, II, III, and V are most frequently associated with invasive disease in North America [3]. Only serotypes Ia and III were found in Thailand [17] while we found three serotypes in the present study. The rapid and unexpected evolution of GBS strains has led to significant differences in recent findings that are of concern to researchers and treatment teams. The genomic differences between the current common type (II and III) and the previously common type (III) are significantly diverse, for which the pathogenic and viral characteristics of the species are very different. GBS colonization is common in Gambian women at delivery and in their infants. The predominant serotypes were reported as V (55%), II (16%), III (10%), Ia (8%), and Ib (8%) [47]. Udo and co-workers [9] detected was lmb, scpB, dltR, cfb, sodA and bca as the most common gene cluster in 13.6% of the GBS isolates. Similarly, Duarte et al. [19] reported the presence of a cluster of genes consisting of bca, lmb and scpB in 66.9% of GBS isolates of human origin and scpB in 44.7% of GBS isolates of bovine origin highlighting the importance of multiple virulence factors to the success of GBS isolates as pathogens. The α- and β-antigens of protein C, encoded by bca and bac, and the Rib protein encoded by rib, have been investigated as possible vaccine candidates because of their ability to elicit protective immunity against GBS infections [4,48]. However, the detection of CylE, lmb, and bca in the isolates of this study suggests that a GBS vaccine targeting these proteins would be effective against our population.
This study has shown that GBS isolates obtained in Kosar hospital belonged to diverse genetic backgrounds with the majority carrying multiple virulence genes. These findings provide a profile for the molecular epidemiology, multilocus sequence typing, antimicrobial resistance, and virulence gene clustering of GBS isolates in Ghazvin and contribute to the body of knowledge on the distribution of virulence-associated genes in GBS in general. The results may be beneficial for the clinical monitoring, prevention, and control of GBS. Due to the moderate to a high prevalence of GBS colonization in pregnant women in Iran, screening of all high-risk pregnant women at 35 to 37 weeks for the presence of GBS, antibiotic susceptibility, common serotypes, and initiatives to produce vaccines for common serotypes are recommended.
The study was approved by the Scientific Ethical Committee of Qazvin University of Medical Sciences. All participants provided written informed consent.
We wish to thank the Vice-Chancellor for Research and Technology Affairs of the Qazvin University of Medical Sciences for supporting this research. Our heartiest thanks and gratitude to Ms. Fahimeh Baloo for the collection of clinical samples.
ZR carried out the experimental laboratory works, analyzed the results, and drafted the manuscript. SSHSA conceptualized the study. HP reviewed the findings and revised the first draft of the manuscript. FB collected the clinical samples. All authors were involved in the preparation of the manuscript.
The authors declare that there are no conflicts of interest.
- 1. Rosen GH, Randis TM, Desai PV, Sapra KJ, Ma B, et al. (2017) Group B Streptococcus and the vaginal microbiota. J Infect Dis 216: 744-751. [Crossref]
- 2. Jackson LA, Hilsdon R, Farley MM, Harrison LH, Reingold AL, et al. (1995) Risk factors for group B streptococcal disease in adults. Ann Intern Med 123: 415-420. [Crossref]
- 3. Raabe VN, Shane AL (2019) Group B Streptococcus (Streptococcus agalactiae). Microbiol Spectr 7: [Crossref]
- 4. Rosenau A, Martins K, Amor S, Gannier F, Lanotte P, et al. (2007) Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes. Infect Immun 75: 1310-1317. [Crossref]
- 5. Verani JR, McGee L, Schrag SJ (2010) Prevention of perinatal group B streptococcal disease revised guidelines from CDC, 2010. MMWR Recomm Rep 59: 1-36. [Crossref]
- 6. Ulett KB, Benjamin WH Jr, Zhuo F, Xiao M, Kong F, et al. (2009) Diversity of group B streptococcus serotypes causing urinary tract infection in adults. J Clin Microbiol 47: 2055-2060. [Crossref]
- 7. Pérez-Moreno MO, Picó-Plana E, Grande-Armas J, Centelles-Serrano MJ, Arasa-Subero M, et al. (2017) Group B streptococcal bacteriuria during pregnancy as a risk factor for maternal intrapartum colonization: a prospective cohort study. J Med Microbiol 66: 454-460. [Crossref]
- 8. Herbert MA, Beveridge CJ, Saunders NJ (2004) Bacterial virulence factors in neonatal sepsis: group B streptococcus. Curr Opin Infect Dis 17: 225-229. [Crossref]
- 9. Udo EE, Boswihi SS, Al-Sweih N (2013) Genotypes and virulence genes in group B streptococcus isolated in the maternity hospital, Kuwait. Med Princ Pract 22: 453-457. [Crossref]
- 10. Bobadilla FJ, Novosak MG, Cortese IJ, Delgado OD, Laczeski ME (2021) Prevalence, serotypes and virulence genes of Streptococcus agalactiae isolated from pregnant women with 35–37 weeks of gestation. BMC Infectious Diseases 21: 73.
- 11. Rajagopal L (2009) Understanding the regulation of Group B Streptococcal virulence factors. Future Microbiol 4: 201-221. [Crossref]
- 12. Wang P, Ma Z, Tong J, Zhao R, Shi W, et al. (2015) Serotype distribution, antimicrobial resistance, and molecular characterization of invasive group B Streptococcus isolates recovered from Chinese neonates. Int J Infect Dis 37: 115-118. [Crossref]
- 13. Seki T, Kimura K, Reid ME, Miyazaki A, Banno H, et al. (2015) High isolation rate of MDR group B streptococci with reduced penicillin susceptibility in Japan. J Antimicrob Chemother 70: 2725-2728. [Crossref]
- 14. Centers for Disease Control and Prevention (CDC) (2007) Perinatal group B streptococcal disease after universal screening recommendations--United States, 2003-2005. MMWR Morb Mortal Wkly Rep 56: 701-705. [Crossref]
- 15. Dmitriev A, Shakleina E, Tkáciková L, Mikula I, Totolian A (2002) Genetic heterogeneity of the pathogenic potentials of human and bovine group B streptococci. Folia Microbiol (Praha) 47: 291-295. [Crossref]
- 16. Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, et al. (2010) A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 80: 212-214. [Crossref]
- 17. Kannika K, Pisuttharachai D, Srisapoome P, Wongtavatchai J, Kondo H, et al. (2017) Molecular serotyping, virulence gene profiling and pathogenicity of Streptococcus agalactiae isolated from tilapia farms in Thailand by multiplex PCR. J Appl Microbiol 122: 1497-1507. [Crossref]
- 18. Bergseng H, Bevanger L, Rygg M, Bergh K (2007) Real-time PCR targeting the sip gene for detection of group B Streptococcus colonization in pregnant women at delivery. J Med Microbiol 56: 223-228. [Crossref]
- 19. Duarte RS, Bellei BC, Miranda OP, Brito MA, Teixeira LM (2005) Distribution of antimicrobial resistance and virulence-related genes among Brazilian group B streptococci recovered from bovine and human sources. Antimicrob Agents Chemother 49: 97-103. [Crossref]
- 20. Manning SD, Ki M, Marrs CF, Kugeler KJ, Borchardt SM, et al. (2006) The frequency of genes encoding three putative group B streptococcal virulence factors among invasive and colonizing isolates. BMC Infect Dis 6: 116. [Crossref]
- 21. Smith TC, Roehl SA, Pillai P, Li S, Marrs CF, et al. (2007) Distribution of novel and previously investigated virulence genes in colonizing and invasive isolates of Streptococcus agalactiae. Epidemiol Infect 135: 1046-1054. [Crossref]
- 22. Sukhnanand S, Dogan B, Ayodele MO, Zadoks RN, Craver MP, et al. (2005) Molecular subtyping and characterization of bovine and human Streptococcus agalactiae isolates. J Clin Microbiol 43: 1177-1186. [Crossref]
- 23. Wayne P (2010) Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20.
- 24. YektaKooshali MH, Hamidi M, Razavi Tousi SMT, Nikokar I (2018) Prevalence of group B streptococcus colonization in Iranian pregnant women: A systematic review and meta-analysis. International Journal of Reproductive BioMedicine 16. [Crossref]
- 25. Tor-Udom S, Tor-Udom P, Hiriote W (2006) The prevalence of streptococcus agalactiae (group B) colonization in pregnant women at Thammasat Hospital. J Med Assoc Thai 89: 411-414. [Crossref]
- 26. Absalan M (2013) Prevalence of Recto-Vaginal Colonization of Group B Streptococcus in Pregnant Women. Journal of Isfahan Medical School 30.
- 27. Bidgani S, Navidifar T, Najafian M, Amin M (2016) Comparison of group B streptococci colonization in vaginal and rectal specimens by culture method and polymerase chain reaction technique. J Chin Med Assoc 79: 141-145. [Crossref]
- 28. Fatemi F, Chamani L, Pakzad P, Zeraati H, Rabbani H, et al. (2009) Colonization rate of group B Streptococcus (GBS) in pregnant women using GBS agar medium. Acta Medica Iranica 25-30.
- 29. Hassanzadeh P, Motamedifar M, Gharaghani MN (2011) Carriage rate of group B streptococci in pregnant women in three teaching hospitals in Shiraz, Iran. Med Princ Pract 20: 277-282. [Crossref]
- 30. Lambiase A, Agangi A, Del Pezzo M, Quaglia F, Testa A, et al. (2012) vitro resistance to macrolides and clindamycin by group B Streptococcus isolated from pregnant and nonpregnant women. Infect Dis Obstet Gynecol 2012: 913603. [Crossref]
- 31. Rabiei S, M Arab, MR YOUSEFI (2006) Epidemiologic pattern of vaginal colonization by group B Streptococcus in pregnant women in Hamadan, Central west of Iran.
- 32. Bennett JE, Dolin R, Blaser MJ (2014) Mandell, douglas, and bennett's principles and practice of infectious diseases. (9th Edn) 2: Elsevier Health Sciences.
- 33. Eren A, Küçükercan M, Oğuzoğlu N, Unal N, Karateke A (2005) The carriage of group B streptococci in Turkish pregnant women and its transmission rate in newborns and serotype distribution. Turk J Pediatr 47: 28-33. [Crossref]
- 34. Turrentine MA, Colicchia L, Hirsch E, Cheng PJ, Tam MT, et al. (2015) Risk of Recurrence of Group B Streptococcus in Subsequent Pregnancies: A Systematic Review and Meta-analysis:[303]. Obstetrics & Gynecology 125: 97S.
- 35. Fazeli Z, Alipour M, Mozafari NA, Nejad YG, Omran OS, et al. (2015) The survey of Streptococcus agalactiae carriage in pregnant women and determination of antibiotics susceptibity pattern in Amol city. Iranian Journal of Medical Microbiology 9: 20-26.
- 36. Shirazi M, Abbariki E, Hafizi A, Shahbazi F, Bandari M, et al. (2014) The prevalence of group B streptococcus colonization in Iranian pregnant women and its subsequent outcome. International journal of fertility & sterility 7: 267.
- 37. El-Kersh TA, Al-Nuaim LA, Kharfy TA, Al-Shammary FJ, Al-Saleh SS, et al. (2002) Detection of genital colonization of group B streptococci during late pregnancy. Saudi Med J 23: 56-61. [Crossref]
- 38. Castor ML, Whitney CG, Como-Sabetti K, Facklam RR, Ferrieri P, et al. (2009) Antibiotic resistance patterns in invasive group B streptococcal isolates. Infect Dis Obstet Gynecol 2008: 727505. [Crossref]
- 39. Yasini M, Moniri R, Ghorbaali Z, Ansaripour L, Movahedinejad M, et al. (2014) Prevalence rate, Antibiotic susceptibility and Colonization risk factors of Group B Streptococcus in genital tract of pregnant women. 57: 676-683.
- 40. Mitchell K, Brou L, Bhat G, Drobek CO, Kramer M, et al. (2013) Group B Streptococcus colonization and higher maternal IL-1beta concentrations are associated with early term births. J Matern Fetal Neonatal Med 26: 56-61. [Crossref]
- 41. Corrêa AB, Silva LG, Pinto Tde C, Oliveira IC, Fernandes FG, et al. (2011) The genetic diversity and phenotypic characterisation of Streptococcus agalactiae isolates from Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 106: 1002-1006. [Crossref]
- 42. Sadeh M, Firouzi R, Derakhshandeh A, Bagher Khalili M, Kong F, et al. (2016) Molecular characterization of Streptococcus agalactiae isolates from pregnant and non-pregnant women at Yazd University Hospital, Iran. Jundishapur J Microbiol 9: e30412. [Crossref]
- 43. Mashouf RY, Mousavi SM, Rabiee S, Alikhani MY, Arabestani MR (2014) Direct identification of Streptococcus agalactiae in vaginal colonization in pregnant women using polymerase chain reaction. Journal of Comprehensive Pediatrics 5.
- 44. Wójkowska-Mach J, Pomorska-Wesołowska M, Romanik M, Romaniszyn D (2021) Prevalence and Antimicrobial Susceptibility Profiles of Microorganisms Associated with Lower Reproductive Tract Infections in Women from Southern Poland—Retrospective Laboratory-Based Study. Int J Environ Res Public Health 18: 335. [Crossref]
- 45. Armistead B, Whidbey C, Iyer LM, Herrero-Foncubierta P, Quach P, et al. (2020) The cyl Genes Reveal the Biosynthetic and Evolutionary Origins of the Group B Streptococcus Hemolytic Lipid, Granadaene. Front Microbiol 10: 3123. [Crossref]
- 46. Melin P, Efstratiou A (2013) Group B streptococcal epidemiology and vaccine needs in developed countries. Vaccine 31: D31-D42. [Crossref]
- 47. Le Doare K, Jarju S, Darboe S, Warburton F, Gorringe A, et al. (2016) Risk factors for Group B Streptococcus colonisation and disease in Gambian women and their infants. J Infect 72: 283-294. [Crossref]
- 48. Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, et al. (2005) Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science 309: 148-150. [Crossref]
Editorial Information
Editor-in-Chief
Hiroshi Miyamoto
University of Rochester Medical Center
Article Type
Research Article
Publication history
Received: March 18, 2021
Accepted: April 14, 2021
Published: April 17, 2021
Copyright
©2021 Seyed Aboutorabi SSJ. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Seyed Aboutorabi SSJ, Rasooli Z, Pakniat H, Baloo F (2021) Molecular characterization of prevalence, serotypes, virulence factors, and antibiotic resistance of Streptococcus agalactiae in a maternity hospital, Iran. Integr Cancer Sci Therap 8: DOI: 10.15761/ICST.1000356.






