Abstract
We previously reported that primary cells derived from rat dental pulp exhibit pluripotency, and that stem cell factors such as Stro-1, Ssea-1, Nanog, and Oct3/4 are expressed in these cells. In addition, downregulation of some factors is observed during lineage differentiation. We hypothesized that stem cell markers including OCT3/4, SOX2, and NANOG would be expressed in human oral tissue-derived primary cells. To further evaluate the expression of pluripotent stem cell factors in primary cells derived from human oral tissues, we quantitatively examined the expression of OCT3/4, SOX2, and NANOG in six primary cell lines derived from human gingival fibroblasts, human periodontal ligament fibroblasts, and human oral keratinocytes. Differential expression of OCT3/4, SOX2, and NANOG was detected in primary cells derived from various types of human oral tissue. Moreover, these cells were found to harbor multiple copies of all three genes. Levels of OCT3/4, SOX2, and NANOG correlated well with each other; in primary cells expressing low levels of OCT3/4 and SOX2, the expression of NANOG also tended to be low. The expression of these factors in oral keratinocytes was higher than that in fibroblasts derived from human gingiva and periodontal ligament. This study indicates that primary cells derived from various types of human oral tissues express pluripotent stem cell factors. The results are expected to expand the future reservoir of potential cell sources that are suitable for cell therapy, replacing more controversial sources currently used.
Key words
gene expression, human oral cells, OCT3/4, pluripotency, stem cell
Introduction
Primary cells derived from rodent dental pulp can differentiate into osteogenic, neurogenic, myogenic, and adipogenic lineages under in vitro induction conditions similar to those used for human bone marrow cells [1–5]. Several pluripotency markers have been identified, including Oct3/4 [6–8], Sox2 [9], and Nanog [10,11]. Previously, we reported that stem cell markers for rodents such as Stro-1, Ssea-1, Nanog, and Oct3/4, are expressed in rat dental pulp-derived cells [1]. Oct3/4, Sox2 and Nanog are transcription factors that are strongly expressed in embryonic stem cells [9–11], and their downregulation correlates with loss of pluripotency and self-renewal [12]. All are considered markers of primitive stem cells [13].
The epithelia consist of several layers of keratinocytes that have different characteristics. In the oral epithelium, oral keratinocyte stem cells reside close to the basal membrane and the vasculature of the lamina propria. These cells possess several attributes that distinguish them from other oral keratinocytes, including self-renewal potential, ability to differentiate into other cell types from the same tissue, quiescence, and long life span [14]. Oral keratinocyte progenitor cells can replace damaged epithelia with similar traits throughout the human body. Progenitor cells have been shown to regenerate the human cornea during transplantation procedures [15]. Although several studies have focused on the in vitro and in vivo behavior of oral fibroblasts following gingival augmentation, data on oral keratinocytes is scarce [16, 17].
Therefore, we hypothesized that OCT3/4, SOX2, and NANOG would be expressed in primary cells derived from human oral tissues. To confirm this, we performed quantitative expression analysis using primary cells derived from various types of human oral tissues, such as human gingival fibroblasts, human periodontal ligament fibroblasts, and human oral keratinocytes.
Materials and methods
Cells and culture
Human gingival fibroblasts (HGnF) were isolated from human gingiva. They were cryopreserved at passage one and delivered frozen (ScienCell Research Laboratories, Inc., Carlsbad, CA, USA). Human periodontal ligament fibroblasts (HPLF) were isolated from human periodontal tissue. They were also cryopreserved at passage one and delivered frozen (ScienCell Research Laboratories, Inc.). Both HGnF and HPLF are characterized by their spindle morphology and were identified based on immunofluorescence with antibodies specific for fibronectin. HGnF and HPLF were cultured in Fibroblast Medium (ScienCell Research Laboratories, Inc.) supplemented with 10% (v/v) fetal calf serum (ScienCell Research Laboratories, Inc.) on Poly-D-Lysine-coated dishes at 37 °C in a humidified atmosphere containing 5% CO2. CloneticsTM Human Periodontal Ligament Fibroblasts (HPdLF) were isolated from normal human periodontal tissue. Cryopreserved HPdLF were shipped at passage three (Lonza Walkersville, Inc., Walkersville, MD, USA). They were isolated from the ligament that fastens the molars to the jaw bone and stained negative for pan-cytokeratin. HPdLF were cultured in Stromal Cell Basal Medium (Lonza Walkersville, Inc.) supplemented with 0.1% (v/v) human fibroblast growth factor-B, 0.1% (v/v) human recombinant insulin, and 5% (v/v) fetal bovine serum (Lonza Walkersville, Inc.) on Poly-D-Lysine-coated dishes at 37 °C in a humidified atmosphere containing 5% CO2. Human oral keratinocytes (HOK) were isolated from human oral mucosa. They were cryopreserved at passage one and delivered frozen (ScienCell Research Laboratories, Inc.). HOK were cultured in Oral Keratinocyte Medium (ScienCell Research Laboratories, Inc.) supplemented with 1% (v/v) Oral Keratinocyte Growth Supplement (ScienCell Research Laboratories, Inc.) on poly-D-Lysine-coated dishes at 37 °C in a humidified atmosphere containing 5% CO2.
RNA extraction
Total RNA was extracted using a QIA shredder and RNeasy Mini Kit (QIAGEN Inc., Germantown, MD, USA) in accordance with the manufacturer’s instructions. The extracted RNA was dissolved in UltraPure DNase/RNase-free distilled water (Life Technologies, Carlsbad, CA, USA), and the concentration of total RNA was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Experiment numbers C3 and C8 were assigned to total extracted RNA from different HGnF clones . C6 and C9 were assigned to total extracted RNA from HPLF and HPdLF, respectively. C1 and C11 were assigned to total extracted RNA from different HOK clones.
Quantitative real-time RT-PCR assay
cDNA was synthesized from total RNA using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA, USA) in accordance with the manufacturer’s instructions. λ polyA+ RNA-A was added as an external reference during the reverse transcription process. As a reference to allow comparison between samples, 2.0 × 108 copies of λ polyA+ RNA-A per 20 µL (Takara Bio Inc., Kusatsu, Shiga, Japan), diluted in EASY Dilution (Takara Bio Inc.), were added per 1 mg of total RNA during the cDNA synthesis process. Real-time RT-PCR was performed using the Fast SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) to examine the expression of each marker. Specific PCR primer sets were designed using the Perfect Real Time Support System (Takara Bio Inc.). The primer sets were as follows: 5´-GCAATTTGCCAAGCTCCTGAA-3´ and 5´- GCAGATGGTCGTTTGGCTGA-3´ for OCT3/4; 5´- GTGAGCGCCCTGCAGTACAA-3´ and 5´-GCTGCGAGTAGGACATGCTGTAG-3´ for SOX2; 5´- TCCAACATCCTGAACCTCAGCTA-3´ and 5´-AGGTTCCCAGTCGGGTTCAC-3´ for NANOG. PCR amplification was performed using the StepOne Plus system (Applied Biosystems) with an initial denaturation step of 20 sec at 95 °C, followed by 40 cycles each of 95 °C for 3 sec and 60 °C for 30 sec, finishing with an automatic melting curve stage. The values were adjusted using real-time primers for λ polyA (Takara Bio Inc.) as an internal control. The expression levels are presented as the fold change, comparing values after induction to those before induction, and were calculated using the ∆∆Ct method.
Estimation of expressed copy number
λ polyA+ RNA-A was diluted stepwise to a concentration of 1:10 using EASY Dilution (Takara Bio Inc). A quantitative standard curve was created using 1 × 101 to 1 × 106 copies of diluted λ polyA+ RNA-A. Real-time primers for λ polyA (Takara Bio Inc.) were used for quantitative real-time RT-PCR, which was performed using the Fast SYBR Green PCR Master Mix (Applied Biosystems).
Results
Detection of pluripotent stem cell factors in human oral tissue-derived primary cells
The expression of OCT3/4, SOX2 and NANOG was detected in two HGnF primary cell lines, two primary cell lines derived from human periodontal ligament fibroblasts, and two HOK primary cell lines. The differential expression levels are shown in Figure 1A, B, C.
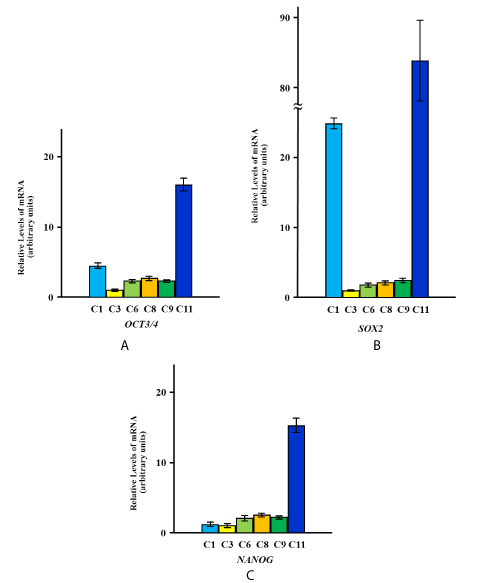
Figure 1. Quantitation of expression levels of OCT3/4, SOX2, and NANOG in primary cells derived from various types of oral tissues.
Expression levels of OCT3/4 (A), SOX2 (B), and NANOG (C) in primary cells derived from various types of oral tissues are shown. The yellow, green, and blue columns represent primary cells from human gingival fibroblasts (C3, C8), human periodontal ligament fibroblasts (C6, C9), and human oral keratinocytes (C1, C11), respectively. The relative mRNA expression levels are presented as the fold change in each cell vs. C3 as a reference, and represent the mean values of six independent assays for each cDNA sample. The values were calculated using Relative Quantity (RQ) Study Software and show statistical variability in terms of the calculation of each sample’s RQ value. The RQmin/RQmax values are graphically represented as error bars.
Copy numbers of pluripotent stem cell factors in oral tissue-derived primary cells
Standards consisting of 101, 102, 103, 104, 105, and 106 copies had Ct values with mean ± standard deviation (SD) of 35.1 ± 0.52, 30.8 ± 0.13, 27.5 ± 0.03, 24.0 ± 0.01, 20.9 ± 0.02, and 17.5 ± 0.06, respectively. A standard curve was produced to directly compare copy number to Ct values for a copy number range of 1 × 101 to 1 × 106 (Figure 2). The mean Ct value was dependent on the serial dilution of λ polyA+ RNA-A.
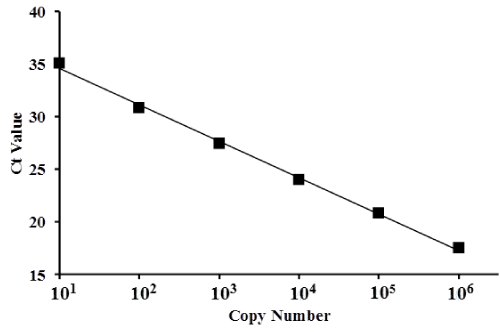
Figure 2. Scatter plot and standard curve of gene copy numbers based on λ polyA+ RNA-A.
Ct values (represented by squares) were plotted for serially diluted samples of λ polyA+ RNA-A. The x-axis represents copy number and the y-axis represents the Ct value. The standard curve for copy number ranged from 1 × 101 to 1 × 106 (depicted in the graph).
The mean Ct values for OCT3/4, SOX2, and NANOG for each primary cell line (derived from the various types of human oral tissues) are shown in Table 1. The estimated copy number for each marker was plotted on the standard curve. Using the standard curve, the copy numbers for OCT3/4, SOX2, and NANOG in each primary cell line were estimated and these values are presented in Table 2.
Table 1. Mean Ct values of OCT3/4, SOX2, and NANOG in each primary cell line derived
from various types of human oral tissues.
|
C1 |
C3 |
C6 |
C8 |
C9 |
C11 |
OCT3/4 |
31.3 |
33.6 |
32.9 |
32.0 |
32.2 |
29.8 |
SOX2 |
31.1 |
35.8 |
35.4 |
34.5 |
34.3 |
29.6 |
NANOG |
34.3 |
34.3 |
34.6 |
34.1 |
33.1 |
33.3 |
Table 2. Copy numbers of OCT3/4, SOX2, and NANOG in each primary cell line derived
from various types of human oral tissues.
|
C1 |
C3 |
C6 |
C8 |
C9 |
C11 |
OCT3/4 |
90.2 |
19.5 |
31.1 |
56.6 |
49.6 |
245.0 |
SOX2 |
103.1 |
4.5 |
5.9 |
10.7 |
12.2 |
28.0 |
NANOG |
12.2 |
10.0 |
14.0 |
27.2 |
23.8 |
11.8 |
Discussion
Some reports suggest that cells positive for pluripotent stem cell factors such as OCT3/4 and SOX2 might represent reprogrammed cancer stem cells that induce oral carcinogenesis [18]. The expression of OCT3/4, SOX2, NANOG, and long non-coding RNAs, which regulate reprogramming, is associated with the development and prognosis of oral squamous cell carcinoma [19,20]. Side population (SP) cells have been identified as a side branch of Hoechstlow cells that comprise a darker fraction than cells in the G0/G1 phase in flow cytometry, and are present as cancer stem cells in many human cancers and cancer cell lines. We previously reported stem-like properties including pluripotency, self-renewal, and retention of an undifferentiated state in this subpopulation of human oral cancer SSC-4 cells. OCT3/4 and NANOG levels were significantly higher in SP cells than in non-SP cells, suggesting that a subset of oral cancer cells has the potential to act as stem cells [21]. The levels of pluripotent stem cell factors have been well characterized in cancer stem cells; however, they require further characterization in normal cells derived from human oral tissues.
The present study found that OCT3/4, SOX2, and NANOG, pluripotent stem cell factors, are differentially expressed in primary cells derived from various types of oral tissues. Copy numbers of OCT3/4, SOX2, and NANOG in these primary cells ranged from 19.5 to 245.0, 4.5 to 103.1, and 10.0 to 27.2, respectively. In primary cells expressing low levels of OCT3/4 and SOX2, the level of NANOG also tended to be low. Although the expression of pluripotent stem cell factors in oral keratinocytes was relatively higher compared to that in fibroblasts derived from human gingiva and periodontal ligaments, the relationship between stem-like properties such as pluripotency or self-renewal and the expression of these factors remains unclear.
Recently, we reported that the direct conversion of dental pulp cells to cells of developmentally unrelated tissues involves miRNA-dependent regulation [22]. Pluripotent stem cell factors and intercellular signaling factors might be fundamentally regulated in a genetic and epigenetic manner, such that the expression of these factors might affect significant biological functions including stem-like properties. Highly efficient miRNA-mediated reprogramming of somatic cells has previously been reported [23,24]. The expression of the miR-302/367 or miR-302/372 cluster rapidly and efficiently reprograms somatic cells into iPS cells without exogenous transcription factors such as Oct3/4 or Sox2. It is possible that pluripotent stem factors are regulated by intracellular signaling via small RNAs from exosomes, for example. This study has expanded the potential cell sources suitable for regenerative medicine. Indeed, factors secreted by human dental pulp stem cells have been identified to have multifaceted therapeutic benefits for recovery after several types of injuries or diseases, such as those of the spinal cord, heart, and liver [25-27]. This study determined the exact copy numbers of pluripotent stem cell factors in primary oral cells. Therefore, primary cells derived from oral tissues expressing these factors or factors secreted by primary cells represent an attractive and promising tool in the field of tissue regeneration and engineering. We believe that the primary cells derived from various types of human oral tissues will become an acceptable source of cells for cell therapy in the future, replacing the more controversial sources currently used.
Funding information
This study was supported, in part, by a Grant-in-Aid for Scientific Research (21592547, 24593020, 15K11283) from the Japan Society for the Promotion of Science (Nozaki T).
Acknowledgements
This study was performed, in part, at the Institute of Dental Research, Osaka Dental University.
Competing interest
The authors declare that there is no conflict of interest.
References
- Nozaki T, Takeyasu M, Hirao A, Shinohara M, Daito M, et al. (2005) Differentiation of rat dental pulp-derived cells into an osteoblastic lineage. Oral Sci Int 2: 118-125.
- Takeyasu M, Nozaki T, Watanabe M, Shinohara M, Morita J, et al. (2004) In vitro osteogenic differentiation potential of dental pulp stem cells. J Oral Tissue Engin 2: 25-30.
- Takeyasu M, Nozaki T, Daito M (2006) Differentiation of dental pulp stem cells into a neural lineage. Pediat Dent J 16: 154-162.
- Nakatsuka R, Nozaki T, Uemura Y, Matsuoka Y, Sasaki Y, et al. (2010) 5-Aza-2'-deoxycytidine treatment induces skeletal myogenic differentiation of mouse dental pulp stem cells. Arch Oral Biol 55: 350-357. [Crossref]
- Nozaki T, Ohura K (2011) Gene expression profile of dental pulp cells during differentiation into an adipocyte lineage. J Pharmacol Sci 115: 354-363. [Crossref]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, et al. (1990) A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345: 686-692. [Crossref]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P (1990) Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J 9: 2185-2195. [Crossref]
- Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P (1990) New type of POU domain in germ line-specific protein Oct-4. Nature 344: 435-439. [Crossref]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, et al. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126-140. [Crossref]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643-655. [Crossref]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, et al. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631-642. [Crossref]
- Carlin R, Davis D, Weiss M, Schultz B, Troyer D (2006) Expression of early transcription factors Oct-4, Sox-2 and Nanog by porcine umbilical cord (PUC) matrix cells. Reprod Biol Endocrinol 4: 8. [Crossref]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947-956. [Crossref]
- Rusu D, Calenic B, Greabu M, Kralev A, Boariu M, et al. (2016) Evaluation of oral keratinocyte progenitor and T-lymphocyte cells response during early healing after augmentation of keratinized gingiva with a 3D collagen matrix - a pilot study. BMC Oral Health 17: 9. [Crossref]
- Chen HC, Chen HL, Lai JY, Chen CC, Tsai YJ, et al. (2009) Persistence of transplanted oral mucosal epithelial cells in human cornea. Invest Ophthalmol Vis Sci 50: 4660-4668. [Crossref]
- Nocini PF, Zanotti G, Castellani R, Grasso S, Cristofaro MG, et al. (2013) Bilayered collagen nano-structured membrane prototype (collagen matrix 10826®) for oral soft tissue regeneration: an “in vitro” study. Clin Oral Implants Res 24: 612-617.
- Willershausen I, Barbeck M, Boehm N, Sader R, Willershausen B, et al. (2014) Non-cross-linked collagen type I/III materials enhance cell proliferation: in vitro and in vivo evidence. J Appl Oral Sci 22: 29-37. [Crossref]
- Cai J, He B, Li X, Sun M, Lam AK, et al. (2016) Regulation of tumorigenesis in oral epithelial cells by defined reprogramming factors Oct4 and Sox2. Oncol Rep 36: 651-658. [Crossref]
- Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Arun K, Vinothkumar V, et al. (2017) Expression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancer. Tumour Biol 39: 1010428317698366. [Crossref]
- Fu TY, Hsieh IC, Cheng JT, Tsai MH, et al. (2016) Association of OCT4, SOX2, and NANOG expression with oral squamous cell carcinoma progression. J Oral Pathol Med 45: 89-95. [Crossref]
- Nozaki T, Nishiitsutsuji R, Ohura K (2016) Stem cell-like molecular and morphological properties in a subpopulation of human oral cancer cells. J Oral Biol 3: 4.
- Nozaki T, Ohura K (2017) Inhibition of miR-183 induces insulin in dental pulp cells. J Hard Tissue Biol 26: 319-322.
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, et al. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376-388. [Crossref]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, et al. (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29: 443-448. [Crossref]
- Yamamoto A, Sakai K, Matsubara K, Kano F, Ueda M (2014) Multifaceted neuro-regenerative activities of human dental pulp stem cells for functional recovery after spinal cord injury. Neurosci Res 78: 16-20. [Crossref]
- Yamaguchi S, Shibata R, Yamamoto N, Nishikawa M, Hibi H, et al. (2015) Dental-pulp-derived stem cell conditioned medium reduces cardiac injury following ischemia-reperfusion. Sci Rep 5: 19295. [Crossref]
- Hirata M, Ishigami M, Matsushita Y, Ito T, Hattori H, et al. (2016) Multifaceted therapeutic benefits of factors derived from dental pulp stem cells for mouse liver fibrosis. Stem Cells Transl Med 5: 1416-1424. [Crossref]


