Identifying viral-host interactions is central in understanding the pathogenesis of intracellular agents, such as bovine viral diarrhea viruses (BVDVs). A high expression level of BVDV NS3 protein is associated with its cytopathic (CP) biotype and is essential in viral replication and viral-induced apoptosis. Although the differences between CP and non-cytopathic BVDV biotypes are well established, especially the importance of the BVDV NS3 viral protein in giving rise to different biotypes, only a few studies identified BVDV-cellular partners. In this study, using a proteomics approach, we identified 71 bovine proteins that interact with the CP NS3 protein at three different time-points post-infection. Our results show that BVDV-host interaction dynamic network involves simultaneously and sequentially interacting proteins in different compartments of the host cells. System analysis of targeted proteins shows that CP BVDVs manipulate multiple and different pathways, primarily the ribosome/translation pathway, specifically at early stages of infection. At later stages of infection, when high levels of NS3 are present, apoptosis can be detected in infected cells. In addition, combining results from this study with previously identified protein interactions adds an extra dimension and higher connectivity to the BVDV strain NADL-host interaction network. Overall, our results indicate correlations among an increase in viral RNA translation, free NS3 level, and cytopathic effect associated with CP biotype infection. Finally, our approach highlights the dynamic interplay between BVDVs and host cells and identifies specific CP BVDV-host interactions that can be used as specific targets for further investigation as BVDV antiviral therapies.
bovine viral diarrhea, pestivirus
Abbreviations
BVDV: Bovine Viral Diarrhea Virus; CP: Cytopathic; NCP: Noncytopathic; HCV: Hepatitis C Virus; BT: Bovine turbinate; MS: Mass spectrometry; ATCC: American Tissue Culture Collection; FBS: Fetal Bovine Serum; ANOVA: Analysis of Variance; IP: Immunoprecipitation; ACN: Acetonitrile; HPLC: High Performance Liquid Chromatography; GO: Gene Ontology; SEA: Singular Enrichment Analysis; SEACOMPARE: Cross Comparison of SEA; ROS: Reactive oxygen species; ∆ᴪm: Mitochondrial membrane potential.
Bovine viral diarrhea viruses (BVDVs), single-stranded RNA viruses in the Pestivirus genus within the Flaviviridae family, are significant disease-causing agents having a major economic impact on the cattle industry, causing both productive and reproductive losses [1-3]. Unlike other Flaviviridae family members, BVDVs exist as two biotypes, cytopathic (CP) or noncytopathic (NCP). One of the most striking biochemical differences between BVDV biotypes is in the processing differences of the BVDV NS2-3, a 120kD protein that can be cleaved into NS2 (40kD) and NS3 (80kD) by autoprotease activity of NS2 [4,5]. Free NS3 produced by NS2-3 cleavage is generated continuously following CP infections, whereas limited cleavage of NS2-3 to release NS3 occurs at early stages of NCP infection to promote genome translation and replication. It was demonstrated that NS3 of viruses of Flaviviridae has serine-protease activity at the N-terminus and RNA-helicase activity at the C-terminus [6,7]. Current research indicates that the induction of a cytopathic effect with CP infection correlates with a dramatic increase of viral RNA synthesis through the release of free NS3 [8] and with the N-terminal serine protease domain activity of NS3 [9]. Therefore, developing effective therapies against BVDV is dependent on identifying cellular targets of viral proteins, specifically NS3, and on the consequences of those interactions on cellular signalling pathways. Few studies have identified BVDV-host protein-protein interactions [4,10-12], and of those studies one showed sphingosine kinase 1 (SPHK1) as a CP NS3 interacting partner [10].
Studies demonstrate that RNA viruses manipulate multiple host pathways in order to evade the immune response, control cell survival, hijack protein translation, affect cytoskeleton remodelling, and alter cytoplasmic transport [13-16]. Recently, several high-throughput studies using multiple Flaviviridae viruses identified and analysed virus-host interactions networks that highlighted the role of different viral proteins in the pathogenesis of associated diseases [15,17]. For example, analysis of hepatitis C virus (HCV, a Flaviviridae virus closely related to BVDV) infection network showed that NS3, with 214 cellular partners, is the most connected viral protein [17]. Enrichment analysis showed pathways associated with HCV clinical syndromes and identified focal adhesion as a novel pathway affected mainly by HCV NS3 and NS5A proteins. However, these results were obtained at a single time-point post infection. Because interactions happen not only simultaneously but also sequentially with time, in this study we used a proteomics-based approach to monitor the dynamics of BVDV-host interactions at multiple stages of the viral life cycles. NS3-host complexes were co-immunoprecipitated from CP BVDV-infected and uninfected bovine turbinate (BT) cells using NS3-specific monoclonal antibody, and co-purified cellular proteins were identified by mass spectrometry (MS). This study identified 71 cellular proteins interacting with BVDV NS3 and that NS3 interacted with different host proteins with similar or differing biological functions at each stage of infection. Systems biology analysis of NS3-interacting host proteins showed enhanced viral RNA translation and activation of cellular pathways inducing apoptosis, functions activated by the CP but not by the NCP biotype. Taken together, this time-dependent study, which is the first comprehensive evaluation of host proteins targeted by the CP BVDV NS3 viral protein, reveals that cellular changes depend on the BVDV replication cycle. Our results highlight the importance of viral RNA accumulation at an early stage of infection that activates cellular pathways triggering apoptosis that contribute to the cytopathic effect induced by BVDV. Our data support our previous findings that increased NS3 expression correlates with apoptosis in CP-infected cells, a process not seen during NCP infection when the NS23 product is not effectively cleaved to produce NS3 [18].
Cell line and BVDV infection
BVDV-free BT cell line was obtained from the American Tissue Culture Collection (ATCC) and propagated in Dulbecco minimal essential media (DMEM, Invitrogen) supplemented with 2 mM L-glutamine, 1.5 g/l sodium bicarbonate, 50 µg/ml gentamicin, and 10% fetal bovine serum (FBS) at 37°C in 5% CO2. BT cell monolayers then were infected with CP BVDV NADL strain (ATCC) at a multiplicity of infection (MOI) of 0.2 and sampled at the indicated time-points post-infection. Uninfected BT cells are used as a negative control.
Flow cytometry
To assess NS3 expression levels using flow cytometry, cells from each time-point post-infection of either CP BVDV-infected or uninfected samples were first trypsinized (TrypLE™ Express, life technology) for 10 minutes. For intracellular staining, 0.5 × 106 cells were fixed and permeabilized using BD Cytofix /Cytoperm Kit (BD Biosciences) solution for 20 min at 4°C. Cells were stained with 2.5 μg/ml anti-BVDV NS3 mAb (EuroClone) or isotype control IgG for 1 hour at room temperature according to the manufacturer’s protocol. After washing, cells were incubated with FITC-conjugated goat anti-mouse Ig for 1 hour at room temperature in the dark and then analyzed using a FACS Calibur (BD Biosciences). To exclude cellular debris from the analyses, we used a conventional scatter gating method. Resulting data were analysed using FlowJo software (Treestar) as duplicates. Data were analyzed using analysis of variance (ANOVA). Differences with a P-value ≤ 0.05 were considered statistically significant.
Co-immunoprecipitation and Western blot
Cell lysates were prepared from CP BVDV-infected or uninfected cells at each time-point post-infection using immunoprecipitation (IP) lysis/wash buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, pH 7.4; Pierce). For immunoprecipitation, 50 μg cell lysate were incubated in a mixture consisting of 50 μL of 100 μg/mL anti-BVDV NS3 mAbs (EuroClone) and 4 μl 100x protease inhibitor (Pierce) in lysis/wash buffer in a final volume of 400 μL for 4 hours with rotation at 4°C. The mixtures were then added to 50 μL of Dynal magnetic beads washed previously with IP lysis/wash buffer, and incubation continued overnight at 4°C. After incubation, the beads were washed three times with IP lysis/wash buffer, and the precipitated proteins were eluted by incubation in 20 μl of 50 mM glycine, pH 2.8 at 70°C. Samples were then recovered from the beads and neutralized with 1M Tris pH 9.5.
For western blot, co-immunoprecipitated proteins at 42 hours post-infection were resolved on 10% SDS-PAGE and transferred to a nitrocellulose membrane, as described previously [19]. Briefly, membranes were blocked overnight at 4°C in Tris buffered saline (TBS) containing 20% goat serum and 5% BSA. Membranes were then washed and incubated with anti-BVDV NS3 mAb or a mouse IgG isotype control (Sigma) for 2 hours at room temperature. After washing, membranes were incubated with an alkaline phosphatase-conjugated goat anti-mouse Ig (Southern Biotechnology Associates) at room temperature for 1 hour. Protein bands were developed with Sigma Fast NBT/BCIP. Western blotting experiments were done in duplicates.
Mass spectrometry
Protein analysis: Co-immunoprecipitated proteins were digested with trypsin using the In-solution Digestion kit (Thermo). Briefly, immunoprecipitated proteins were incubated with digestion and reducing buffers for 5 min at 95°C. After cooling, samples were incubated overnight at 37°C with activated trypsin. The resulting mixtures were desalted using a peptide macrotrap (Michrom Bioresources, Inc), dried, and re-suspended in 40 µl of 2% acetonitrile (ACN), 0.1% formic acid (FA), and then one half (~50 µg) of each sample was transferred to low retention high performance liquid chromatography (HPLC) vials for analysis using mass spectrometry, with the remainder frozen at -70°C until required.
Peptide mass spectrometry was accomplished using an EASY-nLC (Thermo Scientific) HPLC machine coupled with an LTQ Velos (Thermo Scientific) linear ion trap mass spectrometer. The EASY-nLC was configured for reverse phase chromatography using a Hypersil Gold KAPPA C18 column (Thermo #25005-150065) with a flow rate of 333 nanoliters per minute. Peptides were separated for mass spectrometry analysis using an acetonitrile gradient starting at 2% ACN, 0.1% FA and reaching 50% ACN, 0.1% FA in 120 minutes, followed by a 15 minute wash of 95% ACN, 0.1% FA. Column equilibration was handled automatically using the EASY-nLC. The eluate from the HPLC was fed directly to the LTQ Velos for nanospray ionization followed by ms/ms analysis of detected peptides. The LTQ Velos was configured to perform 1 ms scan followed by 20 ms/ms scans of the 20 most intense collision induced dissociation (CID) peaks repeatedly over the 172 minute duration of each HPLC run. Dynamic exclusion was enabled with duration of 5 minutes, repeat count of 1, and a list length of 500. The collected spectra were subsequently analyzed using the X!tandem [20] search algorithm.
Raw spectral data from the LTQ Velos were converted to mzML format using the msConvert tool from the ProteoWizard software project [21] because X!tandem could not read the Thermo raw format directly. The fasta database used for peptide spectrum matching (target database) was a combination of the Bos taurus RefSeq protein database and the full length BVDV sequences (AAA42854) from the National Center for Biotechnology Information (NCBI). X!tandem was configured to use tryptic cleavage sites with up to two missed cleavages. Precursor and fragment mass tolerance were set to 1000ppm and 500ppm respectively. Four amino acid modifications were included in the database search: single and double oxidation of methionine and both carboxymethylation and carboxyamidomethylation of cysteine. A decoy search was done using a randomized version of the target database with the same search parameters as before. The search results were filtered using the methods described by Rousseeuw and coworkers [22] and Filzmoser and coworkers [23]. A decoy score distribution was created, and each match from the target database was evaluated as a possible outlier and assigned a probability of being correct. Peptides from the target database were accepted if the probability of being correct was 99% or higher. A list of proteins and identified peptides was generated for each experimental sample.
Protein intensities and identification
Protein intensities for evaluating relative amounts between replicates were calculated from peptide spectral intensity. The raw spectral data were converted to the MS1 tab delimited format [24] using the MakeMS2 tool available from the MacCoss laboratory and the University of Washington [25]. The intensities for each peptide elution peak were pulled from its associated MS1 file using the Perl scripting language and summed. For each identified protein, the peptide intensities were combined and organized by experimental replicate. Cellular proteins were identified from all three trials (with P-values ≤ 0.01) for each of the three time-points post CP-infection; 12, 24 and 42 hours. In addition, for each time-point, proteins identified from the control samples were subtracted out to remove background proteins from the CP-infected samples.
Gene Ontology (GO) enrichment analysis
GO enrichment analysis was done using AgriGO [26] to evaluate functional processes for the NS3-targeted host proteins. For each GO term, AgriGO used a statistical analysis to determine the significance over-representation of this term in our list of proteins compared with the complete list of the bovine-annotated proteins. Singular Enrichment Analysis (SEA) with Fisher test was used to analyze proteins in each time point. Cross Comparison of SEA (SEACOMPARE) was used to compare between time-points with a false discovering rate (FDR) < 0.05 considered significantly enriched.
Network modelling
Interactions among the host proteins and associated significantly enriched pathway and topological analysis were identified using STRING 9.05 [27]. STRING database provided a combined association score for interactions from a variety of angles, such as homology, experiments and text mining to represent the degree of confidence for each interaction.
The Host-Pathogen Interaction Database (HPIDB) [28], a resource that integrates dataset of pathogen-host interactions, was used to determine host proteins that are previously known to be targeted by BVDV NADL proteins.
Identification of NS3-associated host proteins
To determine suitable samples for MS, CP BVDV-infected and uninfected cells were collected initially at each time point and assessed for percentage of virus infection and viability. Compared to uninfected cells, CP BVDV-infected cells showed a significant increase in the NS3 expression levels starting 24 hours post-infection (Figure 1A). Because the increase in NS3 expression at 48 hours was associated with less than 50% of viable cells (previously published data [18] compared to uninfected cells, we selected 42 time-point as the last sample for co-immunoprecipitation. Efficiency of immunoprecipitation with anti-NS3 antibody was validated by western blot analysis of retained fractions, which identified the expected 80 kDa NS3 protein in lysates from CP BVDV-infected cells but not from uninfected and noncytopathic (NCP)-infected cells (Figure 1B).
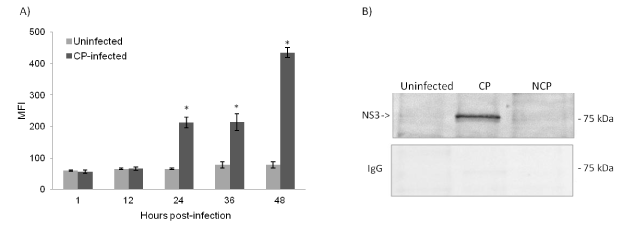
Figure 1. CP BVDV NS3 protein expression (A) Expression of BVDV NS3 in both CP BVDV-infected and uninfected (negative control) cells at the indicated time points are assessed using flow cytometry. NS3 expression levels are shown as mean fluorescence intensity (MFI) and are represented as mean±SD of two independent experiments. The asterisk indicates P-value <0.05 as determined by infected vs. control for each time point. (B) BVDV-infected and uninfected cell lysates at 42 hours post-infection are subjected to immunoprecipitation using an anti-NS3 monoclonal antibody as described in materials and methods. Figure represents one of two experiments.
As a result of our MS analysis, a set of a total of 71 cellular proteins that interacted with NS3 protein was obtained (Supplementary 1). Viral proteins are well known to interact with a large number of host proteins [17,29]. In addition, NS3 of the CP biotype NADL include multiple intrinsically disordered regions (according to DISOPRED2 prediction [30], data not shown) that could aid in interaction with high number of cellular partners. Of the 71 interacting cellular proteins, 26, 37, and 9 proteins were identified at 12, 24 and 42 hours post-infection, respectively. Only one of these identified proteins, the keratin KRT5 protein, was returned at two time-points, 24 and 42 hours, of infection which implies different binding requirements during the viral life cycle. The few interactions at 42 hours could be due to the loss of cell membrane integrity and that the virus is at the release stage.
Functional analysis of cellular proteins associated with different stages of CP infection
Determining the normal cellular functions of host proteins that interact with BVDV is a crucial step for understanding how BVDV subverts host cellular machinery. Subsequently, proteins identified at each time-point were categorized according to their over-represented (FDR < 0.05) GO annotation terms for cellular distributions (Figure 2A) and biological (Figure 2B) and molecular functions (Figure 2C). Comparison of GO terms excluded the 42 hours’ time-point because the low number of host proteins was not eligible for enrichment and comparison.
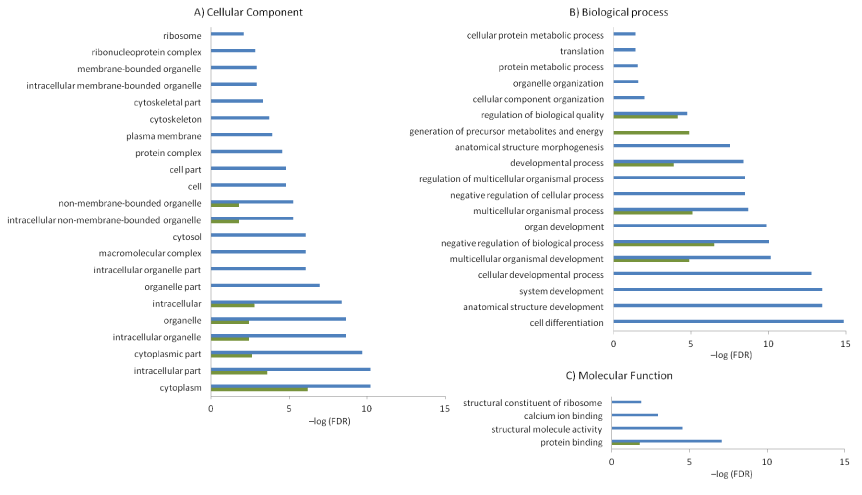
Figure 2. Gene Ontology (GO) functional enrichment Significantly enriched GO terms (FDR < 0.05) in (A) cellular components, (B) biological processes and (C) molecular functions associated with host proteins at 12 (green) and 24 (blue) hours post-infection are identified by AgriGO enrichment analysis. X- and y-axis represent –log (FDR) and GO terms, respectively.
At the earlier stage of BVDV infection (12 hours), our analysis showed that CP NS3-interacting host proteins were mainly associated with GO terms including; negative regulation of cell biological processes, energy generation, and the cytoplasm. To start with, functions that derive energy and all other metabolic functions from the host cell are required by viruses because viruses cannot generate and store energy. This indicates that NS3 manipulated the cell cycle at an early stage.
In addition to sharing most of the same functions as the 12-hour time-point infection, the 24 hour finding showed a higher and wider impact on enriched cellular functions. CP NS3 at 24 hours post-infection interacted with proteins enriched in cell differentiation and development, negative regulation of cellular processes, translation, ribosomal activity, cytoskeleton, complexes formation, and intracellular membrane parts. Such targeted functional and location terms are in line with the life cycle of RNA viruses. Because viral genomic RNA must first be translated, binding with ribosomal subunits is required to ensure efficient synthesis of viral proteins. It is noteworthy that the translation machinery is one of the most often targeted cellular systems by RNA viruses [13]. Additionally, binding with proteins enriched for intracellular membrane parts commensurate with positive-strand RNA viruses assembling their replication and viral assembly complexes at the intracellular membrane. Previous studies showed that BVDV RNA replication was associated with intracellular membranes and required NS3 through NS5B as well as cellular components [31-34]. Targeting such a set of functions plays a critical role in cell survival and increase of cellular stress. Co-opting these pathways by CP leads to negative regulation of cellular machineries and ultimately apoptosis, which could be the case in the 42 hours samples. In our recent study we showed an increase in reactive oxygen species (ROS) and the breakdown of mitochondrial membrane potential (∆ᴪm) in a time-dependent manner that accompanied an increase in apoptotic BT cells infected with CP but not with NCP BVDV [28], supporting our current proteomics findings. A rapidly growing literature suggests that a transient increase in ROS levels results in the activation of various signalling molecules and pathways to regulate responses to cellular oxidative stresses [18].
Network analysis of inter-connected host proteins targeted by CP BVDV
To identify and visualize interactions among host proteins that interact with NS3, we constructed a protein-protein interaction network for the combined set of all host proteins from all time-points (Figure 3A). Out of 71 proteins, 46 were found to be connected with 16, 27, and 4 of the proteins in the network identified at 12, 24, and 42-hour time-points of infection, respectively. These 46 proteins had 129 interactions, suggesting that NS3 was targeting highly connected host proteins. Pathway enrichment for proteins in this network identified ribosome and protein processing in the endoplasmic reticulum enrichment with six (RPS3, RPL3, RPL13A, RPL24, RPL7A, and RPLP0) and five (PDIA4, PDIA6, CALR, CAPN2, and HSP90AB1) proteins from both 12 and 24 hours infection being the centre of the pathways, respectively (Table 1).
Table 1. Topological analysis of host protein networks from figure 3
Network Parameter |
Network |
| |
A |
B |
C |
Number of nodes |
46 |
23 |
85 |
Number of edges |
129 |
23 |
240 |
Average node degree |
5.61 |
2 |
5.65 |
Clustering coefficient |
0.63 |
0.81 |
0.56 |
Enriched pathway |
Ribosome, Protein processing in the ER |
- |
Ribosome |
To understand better the biological effect of BVDV infection, we collected and analysed proteins that were identified previously to interact with all viral proteins of the BVDV strain NADL. Out of the 59 unique host proteins that interacted with BVDV NADL, only 23 proteins were found to interconnect with no significantly enrichment pathway at P-value ≤ 0.05 (Figure 3B). However, when results from this study were combined with previously identified, a network consisting of 85 proteins (59% of the total proteins identified as interacting with NS3) with 240 interactions (Figure 3C) was formed with ribosomal pathway enrichment.
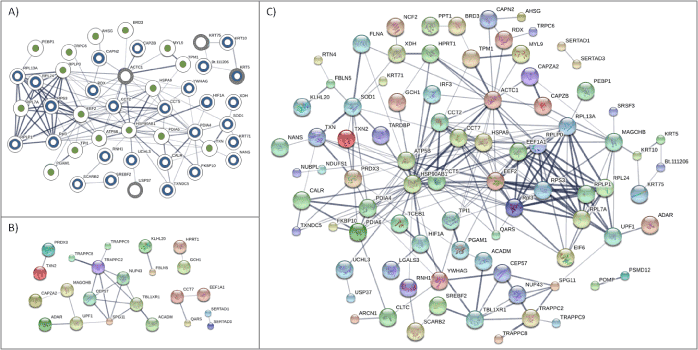
Figure 3. Host protein-protein interaction networks Interaction networks derived from STRING for BVDV strain NADL-targeted cellular proteins (A) identified in this study, (B) previously identified in HPIDB database and (C) a combination of the two data sets. Circles represent host proteins with proteins in panel (A) overlaid with partially coloured circles to represent 12 (green), 24 (blue) and 42 (gray)-hour time-points. Edges represent the interactions between proteins with darker edge signifying higher confidence interactions.
Topology analysis for each of the networks as shown in Figure 3 is summarized in Table 1. Within the three networks, the highest number of interactions was observed in the combined network of host proteins. In addition, topologic analysis indicated that the combined network had the highest connectivity (in term of number of edges, average node degree and clustering coefficient) followed by network from our study and then previously identified proteins. Higher connectivity means that proteins in the network have more interactions among themselves than what would be expected for a random set of proteins of similar size. This shows that incorporating NS3 interactions increases connectivity and size of host protein network targeted by BVDV and indicates common cellular targets during viral infection.
On exposure to a pathogen, multiple interactions with host proteins can occur simultaneously or sequentially in time when specific conditions are fulfilled, bringing a dynamic dimension to pathogen-host interplay. Therefore, identification of specific host proteins targeted by pathogens and their roles during infection contributes to a fundamental understanding of key processes required for infection to occur. Because BVDV, like other RNA viruses, produce relatively few viral proteins, the viral proteins are able to interact with multiple host proteins to subvert different host cell processes during each step of the virus replication cycle. In this study, we report the identification of 71 bovine proteins that associated with BVDV CP NS3 viral protein. However, our study does not eliminate the possibility of other BVDV viral proteins binding with NS3 during precipitation with anti-NS3 antibody. Therefore, the results of this study should be considered as a well-supported hypothesis rather than proven fact and a further confirmation of the cellular protein targeted here with different approaches is needed.
Modelling the roles that targeted host proteins play in BVDV infection and replication shows, not surprisingly, that time-points have unique and overlapping functions, indicating that BVDVs require multiple host cell processes during different stages of viral replication. Because viruses depend on host cells for each step of the viral life cycle, in the earlier stage of infection the NS3 protein begins with utilization the host cell energy pathway. Later, as genome translation occurs soon after internalization and polyprotein uncoating and precedes viral replication [31] the NS3 protein, as expected, bound to cellular proteins that exploit translation machinery. Seizing control of the translational apparatus represents a key event to ensure efficient synthesis of viral mRNA over endogenous host transcripts. We note that targeting ribosomal complexes were shown at both 12 and 24-hour time-points of CP BVDV infection. This strategy allows viral mRNA translation to persist, even under the extreme conditions imposed by the host “shutoff” phenomenon, which severely limits cellular metabolism. Simultaneously, BVDVs manipulate pathways that include negative regulation of cell processes, over-commitment to cellular membranes, complex formation, and cell development and differentiation. The manipulation of these pathways is associated with the increase we detect in NS3 levels and apoptosis in CP-infected cells, which is in agreement with the findings that in CP BVDV-infected cell culture only cells actively synthesizing viral proteins undergo apoptosis [33] and that protease-inactive NS3/4A is unable to induce apoptosis [9]. As a result, we see few binding host proteins and pathways identified later after this stage that emphasize the CP-induced cytopathic effect. In addition, cellular proteins and pathways, such as protein binding, translation, and apoptosis, targeted by CP NS3 are known to be targeted by other viruses, indicating common strategies among these viruses [17,35].
The ability for RNA viruses with few encoded viral proteins (such as BVDVs) to manipulate effectively multiple host pathways is provided by targeting host protein capable of binding multiple proteins in the host protein network. Our study demonstrates this multifunctionality aspect of BVDV viral infection and shows that though different viral stages target different host proteins, those proteins are connected. For example, the centre nodes we identify from host proteins that interact with BVDV NS3 in the translation pathway, RPS3, RPL3, RPL13A, RPL24, RPL7A, and RPLP0, are all members of larger cellular complexes. Similarly, proteins involved in endoplasmic reticulum function, PDIA4, PDIA6, CALR, CAPN2, and HSP90AB1, are highly connected in the network, indicating potential multifunctionality and importance during BVDV replication. These host proteins provide BVDV with the mechanism for co-opting multiple host cell processes during cytopathic infection. Further investigation will elucidate host proteins importance as therapeutic targets. Our study shows also that overall BVDV NADL interactions with its host are focused on targeting the translation and the ribosome activity.
In conclusion, our study reveals new information about BVDV-host interaction and demonstrates the unique dynamic perspective that virally encoded NS3 provides in causing a cytopathic effect. Based on our results, we hypothesize that the cytopathic effect induced by CP BVDV NS3 expression is a by-product of enhanced viral RNA accumulation and activation of cellular pathways triggering apoptosis. Increased cell death can directly contribute to tissue injury and induce inflammation in animals with mucosal disease. Our proteomics results are supported by our recent finding that the time-dependent increase in NS3 expression in BT cells is associated by a time-dependent increase in mitochondrial-induced apoptosis [18]. In addition, our results are well supported by multiple BVDV studies. In particular, Vassilev et al showed that increased levels of RNA accumulation, along with the production of NS3, contribute to the viral cytopathogenicity in cells infected with CP BVDV biotype NADL [8]. Furthermore, Yamane et al showed that dsRNA was the main trigger of apoptosis in CP BVDV-infected cells [36]. In contrast, Schweizer et al demonstrated that NCP BVDV interfered and inhibited dsRNA-induced apoptosis and interferon synthesis [37]. Our host-BVDV interaction models uncover new targets within host that can be targeted either through mutation or molecular inhibition to eliminate infection without sacrificing endogenous host function.
2021 Copyright OAT. All rights reserv
The authors declare that they have no competing interests.
MGA participated in the design of the study, data collection, analysis, drafted the manuscript. GTP participated in the design, assisted in data collection and analysis of immunoassays, helped with manuscript drafting. KP carried out mass spectrometry data generation, participated in protein analysis and manuscript preparation. LMP and FMM initiated the project, participated in the design and coordination of the study, participated in the manuscript drafting, approved final version of the manuscript, provided financial support. All authors contributed to the critical revision of the draft, read and approved the final version of the manuscript.
View supplementary data
- Houe H (2003) Economic impact of BVDV infection in dairies. Biologicals 31: 137-143. [Crossref]
- Houe H (1999) Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol 64: 89-107. [Crossref]
- Kobrak A, Weber EL (1997) [Bovine diarrhea virus: an update]. Rev Argent Microbiol 29: 47-61. [Crossref]
- Rinck G, Birghan C, Harada T, Meyers G, Thiel HJ, et al. (2001) A cellular J-domain protein modulates polyprotein processing and cytopathogenicity of a pestivirus. J Virol 75: 9470-9482.
- Lackner T, Muller A, Pankraz A, Becher P, Thiel HJ, et al. (2004) Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J Virol 78: 10765-10775. [Crossref]
- Warrener P, Collett MS (1995) Pestivirus NS3 (p80) protein possesses RNA helicase activity. J virol 69: 1720-1726. [Crossref]
- Wiskerchen M, Collett MS (1991) Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology 184: 341-350. [Crossref]
- Vassilev VB, Donis RO (2000) Bovine viral diarrhea virus induced apoptosis correlates with increased intracellular viral RNA accumulation. Virus Res 69: 95-107. [Crossref]
- Gamlen T, Richards KH, Mankouri J, Hudson L, McCauley J, et al. (2010) Expression of the NS3 protease of cytopathogenic bovine viral diarrhea virus results in the induction of apoptosis but does not block activation of the beta interferon promoter. J Gen Virol 91: 133-144. [Crossref]
- Yamane D, Zahoor MA, Mohamed YM, Azab W, Kato K, et al. (2009) Inhibition of sphingosine kinase by bovine viral diarrhea virus NS3 is crucial for efficient viral replication and cytopathogenesis. J Biol Chem 284: 13648-13659. [Crossref]
- Johnson CM, Perez DR, French R, Merrick WC, Donis RO (2001) The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J Gen Virol 82: 2935-2943. [Crossref]
- Zahoor MA, Yamane D, Mohamed YM, Suda Y, Kobayashi K, et al. (2010) Bovine viral diarrhea virus non-structural protein 5A interacts with NIK- and IKKbeta-binding protein. J Gen Virol 91: 1939-1948. [Crossref]
- Vidalain PO, Tangy F (2010) Virus-host protein interactions in RNA viruses. Microbes Infect 12: 1134-1143. [Crossref]
- McDermott JE, Diamond DL, Corley C, Rasmussen AL, Katze MG, et al. (2012) Topological analysis of protein co-abundance networks identifies novel host targets important for HCV infection and pathogenesis. BMC Syst Biol 6: 28. [Crossref]
- Le Breton M, Meyniel-Schicklin L, Deloire A, Coutard B, Canard B, et al. (2011) Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol 11: 234. [Crossref]
- Navratil V, de Chassey B, Meyniel L, Pradezynski F, Andre P, et al. (2010) System-level comparison of protein-protein interactions between viruses and the human type I interferon system network. J proteome res 9: 3527-3536. [Crossref]
- de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, et al. (2008) Hepatitis C virus infection protein network. Mol Syst Biol 4: 230. [Crossref]`
- Ammari MG, Pharr GT, Ross MK, Pinchuk GV, Pinchuk LM (2014) Mitochondrial dysfunction associated with viral cytopathogenicity. Current Topics in Virology 2014.
- Wan X, Branton SL, Hughlett MB, Hanson LA, Pharr GT (2004) Expression and Subcellular Location of a Leucine Aminopeptidase of Mycoplasma Gallinarum. International Journal of Poultry Science.
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466-1467. [Crossref]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24: 2534-2536. [Crossref]
- Peter J. Rousseeuw KVD (1998) A Fast Algorithm for the Minimum Covariance Determinant Estimator. Technometrics 41.
- Filzmoser PG, Garrett RG, ClemensR (2006) Multivariate outlier detection in exploration geochemistry. Computers and Geosciences 31.
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, et al. (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom 18: 2162-2168. [Crossref]
- LAB M: http://proteome.gs.washington.edu/software.html.
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64-70. [Crossref]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561-568. [Crossref]
- Ammari MG, Gresham CR, McCarthy FM, Nanduri B (2016) HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford) 2016. [Crossref]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, et al. (2015) Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol cell 57: 349-360. [Crossref]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20: 2138-2139. [Crossref]
- Lindenbach BD, Heinz-Jurgen T, Rice CM (2007) Flaviviridae: The viruses and their replication. In: Fields Virology. (5th edn.) Philadelphia: Lippincott-Raven Publishers, USA.
- Zhang G, Flick-Smith H, McCauley JW (2003) Differences in membrane association and sub-cellular distribution between NS2-3 and NS3 of bovine viral diarrhoea virus. Virus Res 97: 89-102. [Crossref]
- Grummer B, Beer M, Liebler-Tenorio E, Greiser-Wilke I (2001) Localization of viral proteins in cells infected with bovine viral diarrhoea virus. J Gen Virol 82: 2597-2605. [Crossref]
- Son Y, Cheong YK, Kim NH, Chung HT, Kang DG, et al. (2011) Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J Signal Transduct 2011: 792639. [Crossref]
- Wang L, Zhou L, Zhang H, Li Y, Ge X, et al. (2014) Interactome profile of the host cellular proteins and the nonstructural protein 2 of porcine reproductive and respiratory syndrome virus. PloS one 9: e99176. [Crossref]
- Yamane D, Kato K, Tohya Y, Akashi H (2006) The double-stranded RNA-induced apoptosis pathway is involved in the cytopathogenicity of cytopathogenic Bovine viral diarrhea virus. J gen virol 87: 2961-2970. [Crossref]
- Schweizer M, Peterhans E (2001) Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J virol 75: 4692-4698. [Crossref]



