Objective: To explore novel mechanisms for the effect of iguratimod (IGU) on rheumatoid arthritis (RA), we analyzed protein profile changes caused by IGU using chondrosarcoma cells. Methods: OUMS-27 cells were cultured in the presence or absence of 100 μM IGU. Proteins were extracted and separated by 2 dimensional-differential image gel electrophoresis (2D-DIGE). Protein spots of interest were identified by mass spectrometry. Results: 776 and 803 protein spots were detected in the 2D-DIGE results of 24 hour- and 6 day-treatment with IGU, respectively. In the 6 day-treatment, 22 protein spots showed 1.3-fold or higher intensity by IGU-treatment than no-treatment, whereas 15 spots showed -1.3 (1/1.3)-fold or lower intensity (p<0.05). We identified 15 out of the 37 spots, which included proteins involved in packaging and splicing of pre-mRNA, regulation of signaling pathways/protein folding, innate immunity and inflammation, transcription, ATP synthesis, cytoprotection, and cytoskeleton organization. Interestingly, intensity of multiple spots of heterogenous nuclear ribonucleoprotein (hnRNP) A2/B1 and A1 was decreased by the IGU treatment, which are highly expressed in synovia of RA and/or its animal model. Specifically, hnRNP A2/B1 are known as autoantigens in RA and also as proinflammatory activators for NF-kB, the target of IGU. Conclusion: IGU affected protein profiles of chondrosarcoma cells. The decreased expression of hnRNPs suggests novel mechanisms for anti-rheumatic effects of IGU.
chondrosarcoma cells, iguratimod, mass spectrometry, proteomics, rheumatoid arthritis
Rheumatoid arthritis (RA) is a systemic inflammatory disease that causes progressive joint destruction and functional loss [1]. Although its etiology remains unknown, early diagnosis and a treat-to-target approach are recommended as standard care to achieve the remission [2]. Developments of biologics and small-molecule immunosuppressive disease-modifying anti-rheumatic drugs (DMARDs) such as methotrexate and janus kinase inhibitors have enabled this strategy [1, 2]. Nonetheless, it is difficult to use the immunosuppressive drugs in a considerable portion of RA patients with comorbidities such as malignant tumors and chronic infection. Furthermore, for RA patients that achieved the remission, application of treatments with immunomodulatory drugs should be considered.
From these points of view, the role of conventional disease-modifying anti-rheumatic drugs (cDMARDs) is important in RA treatment. The immunomodulatory cDMARDs include salazosulfapyridine and bucillamine which are recommended as alternatives for methotrexate- intolerant patients by RA treatment guidelines [3, 4]. The immunomodulatory cDMARDs can be used in the patients with malignancy and infection. Iguratimod (IGU), a novel immunomodulatory cDMARD developed in Japan, inhibits the production of immunoglobulins and proinflammatory cytokines such as TNFa, IL-1b, and IL-6 [5-8]. IGU has been considered to exhibit its anti-rheumatic effects through suppression of NF-kB, by the inhibition of phosphorylation and nuclear translocation of RelA/p65 without interfering with IkB [6,7,9]. IGU also suppresses ERK and NF-kB pathways in RANKL-induced osteoclast differentiation [9-11]. Recently, IGU was found to block IL-17 signaling in a murine arthritis model, targeting an IL-17 receptor adaptor protein, Act-1 [12]. However, mechanisms of the effects of IGU, especially those on chondrocytes, are still obscured.
In this study, we comprehensively analyzed changes of protein expression of chondrosarcoma cells caused by IGU. We found that IGU affected the protein profile of chondrosarcoma cells. Interestingly, IGU suppressed expression of heterogenous nuclear protein (hnRNP) A2/B1 and A1. Specifically, hnRNP A2/B1 play roles as autoantigens and proinflammatory regulators in RA and also as coactivators for NF-kB, the target of IGU. Our findings suggested novel mechanisms of the effect of IGU on RA treatment.
Cell culture
A chondrosarcoma cell line, OUMS-27 (C-2012-1583, Human Science Research Resources Bank, Sennan, Osaka, Japan) [13], was cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA) and 10% FBS (Hyclone, South Logan, Utah, USA) in type I collagen-coated dishes (AGC techno glass, Haibara-gun, Shizuoka, Japan). To analyze effects of IGU (Tokyo chemical industry, Chuo-ku, Tokyo, Japan) on the cell viability and growth, 1.5x105 of OUMS-27 cells were seeded and cultured for 24 hours in the presence or absence of 10-300 μM of IGU. Similarly, to analyze a longer term (6 days) effect, 1.5x105 of the cells were seeded and 100 μM IGU was added three times every 48 hours. The cells were harvested after 48 hours from the last treatment. All the experiments were performed in triplicates.
Two dimensional-differential image gel electrophoresis (2D-DIGE)
Proteins, extracted from the OUMS-27 cells, were analyzed by 2D-DIGE as previously described [14]. Briefly, an equal weight of proteins taken from six lysates from the 24 hour-cultured cell samples (three of 100 μM IGU-treated and three non-treated) were mixed and labeled with Cyanine dye 3 (Cy3, Cy Dye DIGE Saturation dye, GE Healthcare, Piscataway, NJ, USA) for preparation of an internal control “standard sample”. Each of the six samples was labeled with Cyanine dye 5 (Cy5, GE Healthcare). 2.5 mg of the individual Cy5-labeled samples were mixed with 2.5 mg of the Cy3-labeled standard sample. Then each of the mixed protein sample was applied to an isoelectric focusing (IEF) gel strip (Immobiline Drystrip pH 3-11, 24 cm, non-linear, GE Healthcare). After the proteins were separated by IEF, they were further separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The resultant protein spots were scanned using an image analyzer (Typhoon 9400 Imager, GE Healthcare). The Cy5-fluorescent intensity of each protein spot was normalized by the Cy3-fluorescent intensity of an identical spot by using a quantitative analysis program (Progenesis, Nonlinear Dynamics, Newcastle, UK). The normalized Cy5-intensity was compared between the IGU-treated and non-treated protein samples using Progenesis. Similarly, protein profiles of the 6 day-cultured cells were compared between the IGU-treated and non-treated OUMS-27 cells.
Identification of proteins
Proteins were identified by mass spectrometry (MS) [14]. In brief, 50 mg of proteins was separated by 2-dimensional electrophoresis. Gel specimens corresponding to protein spots of interest were recovered. Then proteins in the gel fragments were digested with trypsin. Peptides produced by the digestion were analyzed using a matrix-assisted laser desorption/ionization time of flight mass spectrometer (MALDI-TOF/MS) (Ultraflex, Bruker Daltonics, Ettlingen, Germany). Based on the mass spectra, some peptides were selected for MS/MS analysis. The obtained MS and MS/MS spectra were used to identify the proteins by database searching (Mascot, http://www.matrixscience.com) against the Swiss Prot human protein sequence database. Protein identification was accepted when MASCOT search results delivered significant MOWSE scores (p<0.05).
Analysis of protein interaction
Interactions among the identified proteins were searched by Ingenuity Pathway Analysis (IPA) (Qiagen, Venlo, NLD). Core analysis was performed using names and the fold differences (IGU/Control) of all the identified proteins and information of all the cells in all the species in IPA database.
Statistical analysis
Significance of differences of cell viability, cell growth, and protein spot intensity in 2D-DIGE was calculated by Student's t-test.
Effects of 24 hour-treatment by IGU on OUMS-27 cells
We first treated OUMS-27 cells for 24 hours with 0-300 mM IGU to examine its effect on viability and growth of the cells. OUMS-27 cells showed viability of more than 94% in the presence of 10-300 μM IGU as well as in the absence of IGU (100%). The cell growth in all the conditions was also at similar levels (0.95- to 1.05-fold) except that in the presence of 300 μM iguratimod (0.84-fold), which showed slightly lower level (p<0.05). Thus, we determined the concentration of IGU as 100 μM and analyzed protein profile of OUMS-27 cells at this condition.
We compared protein profiles of OUMS-27 cells between the 100 μM IGU-treatment and no-treatment by 2D-DIGE. We found 776 protein spots in total (Figure 1A, B). In the 776 protein spots, 41 protein spots showed different intensity between the two conditions (p<0.05) (Table 1). Among the 41 protein spots, 11 protein spots showed 1.3-fold or higher intensity in the IGU-treated cells compared to the non-treated cells. In contrast, 5 spots showed -1.3 (1/1.3)-fold or lower intensity in the IGU-treated cells compared to the non-treated cells. It was shown that IGU affected the protein profile of OUMS-27 cells.
Effects of 6 day-treatment by IGU on OUMS-27 cells
Next, we treated OUMS-27 cells for a longer term of 6 days with 100 μM IGU. The levels of cell viability and growth were high enough and at similar levels at 6 days between the IGU-treated and non-treated cells (100 μM; cell viability, 100.1 %, p=0.92; cell growth, 1.05-fold, p=0.64). Thus, we performed 2D-DIGE using those cell lysates.
In total, 803 protein spots were detected in the 2D-DIGE results (Figure 1C, D). In the 803 protein spots, 150 protein spots showed different intensity between the IGU-treated and non-treated cells (p<0.05, Table 1). Among the 150 protein spots, 22 protein spots showed 1.3-fold or higher intensity in the IGU-treated cells compared to the non-treated cells. On the other hand, 15 spots showed -1.3-fold or lower intensity in the IGU-treated cells compared to the non-treated cells. Protein spots that changed to ±1.3-fold or more in the 6-day treatment (150 spots) showed 3.7-fold in number compared to those in the 24-hour treatment (41 spots) (Table 1).
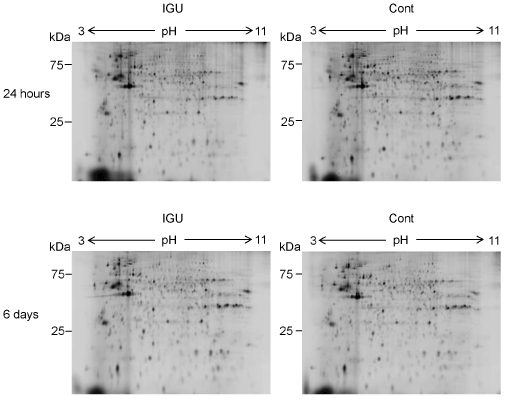
Figure 1. Protein profiles of OUMS-27 cells treated with IGU. Representative results of 2D-DIGE of proteins from OUMS-27 cells cultured in the presence (left upper) or absence (right upper) of 100 μM IGU for 24 hours are shown. Similarly, those cultured in the presence (left lower) or absence (right lower) of 100 μM IGU for 6 days are shown.
Table 1. Change of the OUMS-27 protein profile by the treatment with IGU
Culture periods |
Total number of detected
protein spots |
Number of protein spots
with different intensity
(p<0.05) |
Fold difference, IGU/Control |
Number of the
protein spots |
24 hours |
776 |
41 |
x ≥ 1.5 1.5 > x ≥ 1.3 1.3 > x ≥ -1.3 -1.3 > x ≥ -1.5 -1.5 ≥ x |
2 9 25 3 2 |
6 days |
803 |
150 |
x ≥ 1.5 1.5 > x ≥ 1.3 1.3 > x ≥ -1.3 -1.3 > x ≥ -1.5 -1.5 ≥ x |
7 15 113 11 4 |
To follow a time course of the protein expression, we observed intensity change of the protein spots that showed ±1.3-fold or more change by the 24 hour-treatment with IGU. In the case of 9 intensity-increased spots, intensity of almost all the spots were decreased in the 6 day-treatment (Figure 2A). Similarly, in the case of 4 intensity-decreased spots, intensity of 3 spots were increased in the 6 day-treatment (Figure 2B). The effect of the 24 hour-treatment on OUMS-27 cells was tentative. In contrast, when we observed intensity of the protein spots that showed ±1.3-fold or more change in the 6 day-treatment, almost all the spots of the 22 intensity-increased spots and the 15 intensity-decreased spots similarly increased and decreased in the 24 hour-treatment, respectively (Figure 2C, D). The 6 day-treatment was considered to reflect a long term effect of IGU on OUMS-27 cells.
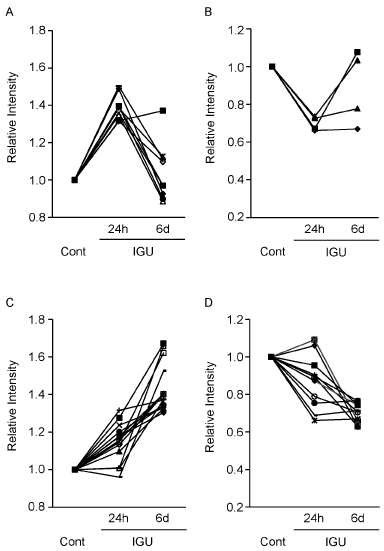
Figure 2. Time course of each protein expression of OUMS-27 cells in the presence of IGU. A, B. Nine out of the 11 protein spots and 4 out of the 5 protein spots, intensity of which were increased to 1.3-fold or more (A) and decreased to -1.3-fold or less (B) in the 24 hour-treatment, respectively (Table 1), were detected both in the 24 hour- and 6 day-treatments and applied for time course analysis. C, D. 14 out of the 22 protein spots and 11 out of the 15 protein spots, intensity of which were increased to 1.3-fold or more (C) and decreased to -1.3-fold or less (D) in the 6 day-treatment, respectively (Table 1), were detected both in the 24 hour- and 6 day-treatments and applied for time course analysis. Control samples (Cont) were proteins from OUMS-27 cells cultured without IGU for 24 hours (A, B) and 6 days (C, D).
Identification of the proteins expression of which was changed by IGU
We tried to identify the proteins in the spots intensity of which was changed by the 6 day-treatment with IGU. 37 protein spots that showed ±1.3-fold or more intensity change by the treatment were subjected to the identification. As a result, 14 proteins were identified from 15 out of the 37 spots (Figure 3, 4, Table 2).
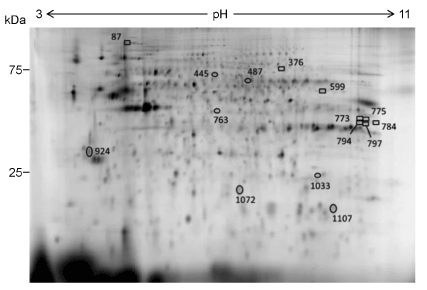
Figure 3. Locations of the identified protein spots on a 2D-DIGE gel. Proteins included in 15 spots out of the 37 spots that showed ±1.3-fold or more change in intensity by the 6 day-treatment with IGU were identified. A representative result of the standard sample is shown.
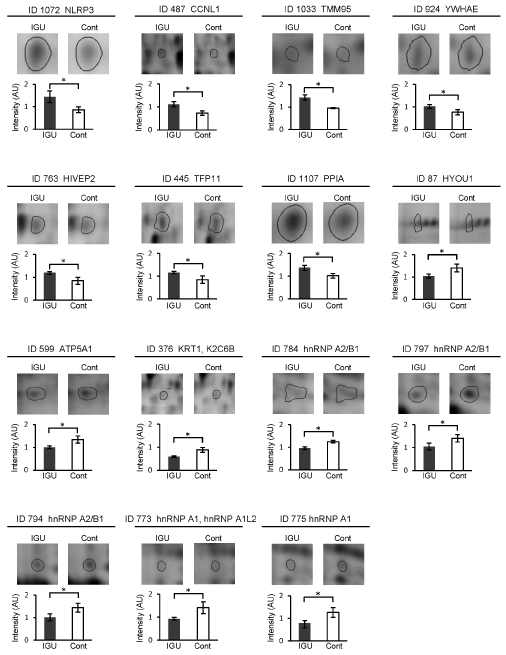
Figure 4. Comparisons of spot intensity of the identified proteins between IGU-treated and non-treated conditions. Representative results of the identified 15 protein spots and intensity difference of those spots between the IGU-treated and non-treated (control) cells are shown. Error bars, standard deviations, *p<0.05.
Table 2. Identification of protein spots, intensity of which was changed by the treatment with IGU
Spot
ID |
Difference (folds) |
Proteins |
Accession ID |
MW (kDa) |
pI |
Mascot
scores |
Coverage (%) |
Confirmed sequences (Mascot ion scores) |
Theoret |
Theoret |
Observed |
Observed |
|
|
|
1072 |
1.67 |
NACHT, LRR and PYD domains-containing protein 3 |
NLRP3_HUMAN |
118.1 |
23 |
6.22 |
6.1 |
71 |
13 |
661MDHMVSSFCIENCHR675 (11) |
487 |
1.53 |
Cyclin-L1 (CCNL1) |
CCNL1_HUMAN |
59.6 |
60 |
10.7 |
6.2 |
75 |
18 |
99LPQVAM*ATGQVLFHR113 (21) |
1033 |
1.50 |
Transmembrane protein 95 (TMM95) |
TMM95_HUMAN |
19.6 |
24 |
8.98 |
7.8 |
57 |
37 |
31LARLCSQMEAR41 (2)
79EAVSSLPSYWSWLRK93 (3) |
763 |
1.40 |
Transcription factor HIVEP2
(HIVEP2) |
ZEP2_HUMAN |
228.9 |
41 |
6.5 |
5.8 |
65 |
8 |
436NALSVTTTSQERAAM*GR452 (11) |
445 |
1.37 |
Tuftelin-interacting protein 11
(TFIP11) |
TFP11_HUMAN |
96.8 |
64 |
5.45 |
5.8 |
61 |
12 |
262ELSQVKVIDMTGR274 (25)
387M*QPDCSNPLTLDECAR402 (7) |
924 |
1.34 |
14-3-3 protein epsilon (YWHAE) |
1433E_HUMAN |
29.2 |
29 |
4.63 |
4.5 |
189 |
18 |
131YLAEFATGNDR141 (54)
131YLAEFATGNDRK142 (16)
154AASDIAMTELPPTHPIR170 (77)
216DSTLIMQLLR225 (18) |
1107 |
1.34 |
Cyclophilin A (PPIA) |
PPIA_HUMAN |
18.0 |
21 |
7.68 |
8.0 |
74 |
10 |
2VNPTVFFDIAVDGEPLGR19 (66) |
87 |
-1.35 |
Hypoxia up-regulated
protein 1 (HYOU1) |
HYOU1_HUMAN |
111.3 |
98 |
5.16 |
5.3 |
66 |
12 |
439DAVVYPILVEFTR451 (24) |
599 |
-1.34 |
ATP synthase subunit alpha,
mitochondrial
(ATP5A1) |
ATPA_HUMAN |
59.7 |
53 |
9.16 |
8.0 |
185 |
11 |
134TGAIVDVPVGEELLGR139 (72)
335EAYPGDVFYLHSR347 (72)
403GIRPAINVGLSVSR416 (15) |
376 |
-1.49 |
Keratin, type II cytoskeletal 1, (KRT1)
Keratin, type II cytoskeletal 6B (K2C6B) |
K2C1_HUMAN
K2C6B_HUMAN |
66.0 66.0 |
69 |
8.15 8.09 |
6.9 |
106 82 |
9 1 |
377YEELQITAGR386 (78)
360YEELQITAGR369 (78) |
784 |
-1.31 |
Heterogenous nuclear ribonucleoproteins A2/B1
(hnRNP A2/B1) |
ROA2_HUMAN |
37.4 |
39 |
8.97 |
9.5 |
99 |
8 |
23LFIGGLSFETTEESLR38 (38)
214GGGGNFGPGPGSNFR228 (46) |
797 |
-1.35 |
Heterogenous nuclear ribonucleoproteins A2/B1
(hnRNP A2/B1) |
ROA2_HUMAN |
37.4 |
38 |
8.97 |
8.8 |
256 |
23 |
23LFIGGLSFETTEESLR38 (52)
154GFGFVTFDDHDPVDK168 (83)
214GGGGNFGPGPGSNFR228 (64) |
794 |
-1.42 |
Heterogenous nuclear ribonucleoproteins A2/B1 (hnRNP A2/B1)
|
ROA2_HUMAN |
37.4 |
38 |
8.97 |
8.7 |
129 |
13 |
23LFIGGLSFETTEESLR38 (47)
214GGGGNFGPGPGSNFR228 (60) |
773 |
-1.52 |
Heterogenous nuclear ribonucleoproteins A1
(hnRNP A1)
Heterogenous nuclear ribonucleoproteins A1-like 2
(hnRNP A1L2) |
ROA1_HUMAN RA1L2_HUMAN |
38.7 34.2 |
40 |
9.17 9.08 |
9.2 |
77 61 |
24
23 |
16LFIGGLSFETTDESLR31 (9)
337SSGPYGGGGQYFAKPR352 (5)
16LFIGGLSFETTDESLR31 (9) |
775 |
-1.60 |
Heterogenous nuclear ribonucleoproteins A1(hnRNP A1)
|
ROA1_HUMAN |
38.7 |
39 |
9.17 |
8.8 |
62 |
8 |
15KLFIGGLSFETTDESLR31(15)
16LFIGGLSFETTDESLR31 (25) |
37 protein spots, intensity of which changed to ±1.3-fold or more by the 6 day-treatment with IGU, were selected for the protein identification. Proteins of 15 out of the 37 spots were identified. MW, molecular weights.
*Oxidation of methionine.
Seven proteins were identified from the 7 spots intensity of which was increased by the treatment with IGU. They were 1) proteins involved in splicing of pre-mRNA (cyclin L1 [CCLN1]; and tuftelin-interacting protein 11 [TFIP11]), 2) proteins that regulate signaling pathways and protein folding (14-3-3 protein epsilon, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon [YWHAE]; and cyclophilin A, peptidyl-prolyl cis-trans isomerase A [PPIA]), 3) a protein involved in innate immunity and inflammation (NACHT, LRR and PYD domains-containing protein 3 [NLRP3]), 4) a transcription factor (human immunodeficiency virus type 1 enhancer-binding protein 2 [HIVEP2]), and 5) a protein with unknown function (transmembrane protein 95 [TMM95]).
The other 7 proteins were included in the 8 spots intensity of which was decreased by the treatment with IGU. They were 1) proteins involved in packaging of pre-mRNA (heterogenous nuclear ribonucleoproteins A2/B1 [hnRNP A2/B1]; heterogenous nuclear ribonucleoproteins A1 [hnRNP A1]; and heterogenous nuclear ribonucleoproteins A1-like 2 [hnRNP A1L2]), 2) a cytoprotective protein from hypoxia (hypoxia up-regulated protein 1 [HYOU1]), 3) an ATP synthase (ATP synthase subunit alpha, mitochondrial [ATP5A1]), and 4) Cytoskeletal proteins (keratin, type II cytoskeletal 1 [KRT1]; and keratin, type II cytoskeletal 6B [K2C6B]).
Interestingly, hnRNP A2/B1 was identified in 3 protein spots (spot ID 784, 794, and 797) (Table 2). Similarly, hnRNP A1 was identified in 2 protein spots (spot ID 773 and 775). In spot ID 773, only hnRNP A1 or both hnRNP A1 and hnRNP A1L2 were considered to be included. Spot ID 376 included KRT1 and/or K2C6B. The observed molecular weights of TFIP11 in the spot ID 445, HIVEP2 in the spot ID 763, and NLRP3 in the spot ID 1072 were smaller than their theoretical weights, indicating that the detected proteins were fragments of those proteins. The pathway analysis showed deduced interactions of 11 out of the 14 identified proteins (Figure 5). hnRNP A2/B1, hnRNP A1 and their complex located at the center of the network. Of note, NF-kB, Akt, and p53 (encoded by TP53 gene) appeared as relay points of the network. These proteins would directly and/or indirectly interact with the identified proteins.
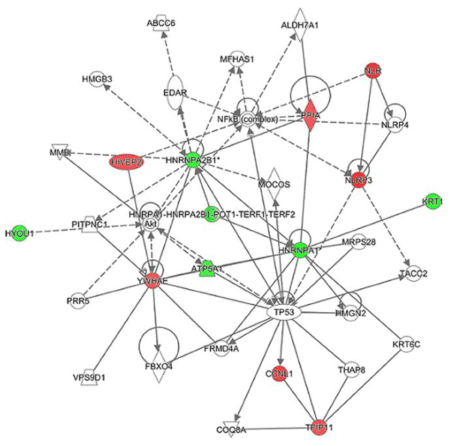
Figure 5. Interaction of the identified proteins. Result of a pathway analysis. Symbols in red and green show the identified proteins intensity of which were increased and decreased by the IGU-treatment, respectively. Solid lines and dotted lines show direct and indirect interactions, respectively. Arrows indicate activation.
2021 Copyright OAT. All rights reserv
We compared protein profiles of a chondrosarcoma cell line OUMS-27 cells between the presence and absence of IGU. Although the maximal plasma concentration after repetitive administration of 50 mg/day IGU for 14 days was 5 μM, the concentration is ununiform in vivo and would reach much higher levels in tissue such as RA-affected joints [15]. Treatment with 100 μM IGU caused significant changes in protein profiles of OUMS-27 cells, that is, intensity of 41 and 150 protein spots was changed by the 24 hour- and 6 day-treatment, respectively (p<0.05) (Table 1). When we examined time courses, intensity of most of the examined spots was much increased or decreased at 6 days compared to that at 24 hours (Figure 2C, D). Continuous treatment with IGU would augment its effect on protein expression.
The 6-day treatment changed intensity of 37 protein spots by 1.3-fold or more (Table 1). Such change levels of protein expression were considered to affect the function of proteins. Among the 14 identified proteins, we focused on three proteins, hnRNP A2/B1, hnRNP A1, and PPIA (Table 2, Figure 4, 5). Of note, intensity of multiple spots of hnRNP A2/B1 and hnRNP A1 was decreased by IGU (Table 2). This indicated that total expression of these proteins would be decreased by IGU regardless of differences of isoforms and/or post-translational modifications.
The main function of hnRNP A/B proteins, components of hnRNP particles, is to package pre-mRNA into hnRNP particles [16]. hnRNP A/B also play roles in telomere maintenance, transcription, pre-mRNA splicing, mRNA nucleo-cytoplasmic export, mRNA stability, and translation. Interestingly, high expression of hnRNP A2/B1 and A1 has been found in inflamed joints of RA patients and/or arthritis models [17-19]. Autoantibodies and autoreactive T cells against hnRNP A2/B1 have been detected in approximately 30% and 60% of RA patients, respectively [17, 20, 21]. Since depletion of hnRNP A2/B1 almost completely blocked development of arthritis both in collagen-induced arthritis (CIA) and K/BxN serum-transfer arthritis, hnRNP A2/B1 was considered to play a crucial role in the pathogenesis of inflammatory arthritis [22]. Besides their role as autoantigens in arthritis, hnRNP A2/B1 act as proinflammatory regulators. hnRNP A2 activates the Raf-MEK-ERK signaling pathway, located at the upstream of NF-kB, by elevating full length A-Raf transcription and reducing production of short dominant-negative isoform of A-Raf [23]. Furthermore, hnRNP A2, phosphorylated and activated by the serine/threonine kinase Akt1, also functions as a transcriptional coactivator for NF-kB c-Rel [24,25]. NF-kB p65 and p50 are known to be targets of IGU [6,7,9,11]. Thus, NF-kB c-Rel activated by hnRNP A2 would be also a target of IGU. Moreover, depletion of hnRNP A2/B1 almost completely inhibited bone erosion in RA models [22]. Since IGU suppresses bone erosion via RANKL- and ERK-mediating pathways [9,11,26,27], its effect to suppress bone erosion may be also associated with the decrease of hnRNP A2/B1.
Our IPA result indicated that both hnRNP A2/B1 and hnRNP A1 could interact with p53 (Figure 5). However, p53 of OUMS-27 cells as well as that of most cancer cells is a mutant-type and thus does not function as a tumor suppressor [13]. Interestingly, suppression of hnRNP A1/A2 induces apoptosis in a variety of human cancer cells irrespective of p53 expression [28]. This was considered because hnRNP A1/A2 have a capping function of telomeres as G-tail binding proteins in cancer cells [28]. Suppression of hnRNPs may inhibit the tumorigenic growth of RA synovial cells.
The other protein of interest, PPIA, was increased by IGU, which has dual function as a chaperone and a signal transducer (Table 2, Figure 4, 5) [29]. Interestingly, PPIA was highly expressed in RA and CIA synovia [30]. PPIA aggravated the arthritis by polarizing macrophages toward proinflammatory M1 phenotypes that produced IL-1β, IL-6, MMP-2, and MMP-9 [30, 31]. The polarization was dependent on the activation of ERK-NF-kB p65 pathway and also on the PPIase activity [30, 32]. On the other hand, in concert with NF-kB p65, PPIA mediates BMP-2-induced Sox9 production and regulation of chondrogenesis signaling [33]. These processes may contribute to repairment of destructed cartilage in RA. Since IGU targets NF-kB, PPIA may be increased to recover the reduced functions of NF-kB.
In conclusion, we found that IGU affected protein profiles of OUMS-27 cells. IGU specifically decreased expression of hnRNP A2/B1 and A1, which are highly expressed in synovia of RA and its arthritis model. Since hnRNP A2/B1 are autoantigens in RA and activators for NF-kB, the decrease of hnRNPs would downregulate the autoimmune responses and also be involved in the NF-kB-targeting mechanism of IGU. The reduction of hnRNP A2/B1 may be further associated with the inhibition of bone erosion by IGU. The present data should be validated using RA chondrocytes and synovial cells in near future. Our results suggest novel mechanisms of the effect of IGU.
The authors indebted Ms Kumi Aso for her technical assistance.
Kimito Kawahata received research grants from Astellas, Eli lilly, Chugai Pharma, and Pfizer, and a personal fee from Ono Pharma.
- McInnes IB, Schett G (2017) Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet 389: 2328-2337. [Crossref]
- Burmester GR, Pope JE (2017) Novel treatment strategies in rheumatoid arthritis. Lancet 389: 2338-2348. [Crossref]
- Lau CS, Chia F, Harrison A, Hsieh TY, Jain R, et al. (2015) APLAR rheumatoid arthritis treatment recommendations. Int J Rheum Dis 18: 685-713. [Crossref]
- Gaujoux-Viala C, Gossec L, Cantagrel A, Dougados M, Fautrel B, et al. (2014) Recommendations of the French Society for Rheumatology for managing rheumatoid arthritis. Joint Bone Spine 81: 287-297. [Crossref]
- Tanaka K, Aikawa Y, Kawasaki H, Asaoka K, Inaba T, et al. (1992) Pharmacological studies on 3-formylamino-7-methylsulfonylamino-6-phenoxy-4H-1-benzopyran-4-one (T-614), a novel anti-inflammatory agent. 4th communication: inhibitory effect on the production of interleukin-1 and interleukin-6. J Pharmacobiodyn 15: 649-655. [Crossref]
- Kohno M, Aikawa Y, Tsubouchi Y, Hashiramoto A, Yamada R, et al. (2001) Inhibitory effect of T-614 on tumor necrosis factor-alpha induced cytokine production and nuclear factor-kappaB activation in cultured human synovial cells. J Rheumatol 28: 2591-2596. [Crossref]
- Aikawa Y, Yamamoto M, Yamamoto T, Morimoto K, Tanaka K (2002) An anti-rheumatic agent T-614 inhibits NF-kappaB activation in LPS- and TNF-alpha-stimulated THP-1 cells without interfering with IkappaBalpha degradation. Inflamm Res 51: 188-194. [Crossref]
- Tanaka K, Yamamoto T, Aikawa Y, Kizawa K, Muramoto K, et al. (2003) Inhibitory effects of an anti-rheumatic agent T-614 on immunoglobulin production by cultured B cells and rheumatoid synovial tissues engrafted into SCID mice. Rheumatology 42: 1365-1371. [Crossref]
- Gan K, Yang L, Xu L, Feng X, Zhang Q, et al. (2016) Iguratimod (T-614) suppresses RANKL-induced osteoclast differentiation and migration in RAW264.7 cells via NF-κB and MAPK pathways. Int Immunopharmacol 35: 294-300. [Crossref]
- Sun Y, Ye DW, Zhang P, Wu YX, Wang BY, et al. (2016) Anti-rheumatic drug iguratimod (T-614) alleviates cancer-induced bone destruction via down-regulating interleukin-6 production in a nuclear factor-κB-dependent manner. J Huazhong Univ Sci Technolog Med Sci 36: 691-699. [Crossref]
- Wei Y, Sun X, Hua M, Tan W, Wang F, et al. (2015) Inhibitory Effect of a Novel Antirheumatic Drug T-614 on the IL-6-Induced RANKL/OPG, IL-17, and MMP-3 Expression in Synovial Fibroblasts from Rheumatoid Arthritis Patients. Biomed Res Int 2015: 214683. [Crossref]
- Luo Q, Sun Y, Liu W, Qian C, Jin B, et al. (2013) A novel disease-modifying antirheumatic drug, iguratimod, ameliorates murine arthritis by blocking IL-17 signaling, distinct from methotrexate and leflunomide. J Immunol 191: 4969-4978. [Crossref]
- Katano M, Kurokawa MS, Matsuo K, Masuko K, Suematsu N, et al. (2017) Phosphoproteome analysis of synoviocytes from patients with rheumatoid arthritis. Int J Rheum Dis 20: 708-721. [Crossref]
- Fiehn C (2010) Methotrexate transport mechanisms: the basis for targeted drug delivery and ß-folate-receptor-specific treatment. Clin Exp Rheumatol 28: S40-45. [Crossref]
- He Y, Smith R (2009) Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci 66: 1239-1256. [Crossref]
- Fritsch R, Eselböck D, Skriner K, Jahn-Schmid B, Scheinecker C, et al. (2002) Characterization of autoreactive T cells to the autoantigens heterogeneous nuclear ribonucleoprotein A2 (RA33) and filaggrin in patients with rheumatoid arthritis. J Immunol 169: 1068-1076. [Crossref]
- Hayer S, Tohidast-Akrad M, Haralambous S, Jahn-Schmid B, Skriner K, et al. (2005) Aberrant expression of the autoantigen heterogeneous nuclear ribonucleoprotein-A2 (RA33) and spontaneous formation of rheumatoid arthritis-associated anti-RA33 autoantibodies in TNF-alpha transgenic mice. J Immunol 175: 8327-8336. [Crossref]
- Hoffmann MH, Tuncel J, Skriner K, Tohidast-Akrad M, Türk B, et al. (2007) The rheumatoid arthritis-associated autoantigen hnRNP-A2 (RA33) is a major stimulator of autoimmunity in rats with pristane-induced arthritis. J Immunol 179: 7568-7576. [Crossref]
- Nell VP, Machold KP, Stamm TA, Eberl G, Heinzl H, et al. (2005) Autoantibody profiling as early diagnostic and prognostic tool for rheumatoid arthritis. Ann Rheum Dis 64: 1731-1736. [Crossref]
- Steiner G (2007) Auto-antibodies and autoreactive T-cells in rheumatoid arthritis: pathogenetic players and diagnostic tools. Clin Rev Allergy Immunol 32: 23-36. [Crossref]
- Herman S, Fischer A, Presumey J, Hoffmann M, Koenders MI, et al. (2015) Inhibition of inflammation and bone erosion by RNA interference-mediated silencing of heterogeneous nuclear RNP A2/B1 in two experimental models of rheumatoid arthritis. Arthritis Rheumatol 67: 2536-2546. [Crossref]
- Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, et al. (2014) Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 20: 505-515. [Crossref]
- Guha M, Tang W, Sondheimer N, Avadhani NG (2010) Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets. Biochim Biophys Acta 1797: 1055-1065. [Crossref]
- Guha M, Fang JK, Monks R, Birnbaum MJ, Avadhani NG (2010) Activation of Akt is essential for the propagation of mitochondrial respiratory stress signaling and activation of the transcriptional coactivator heterogeneous ribonucleoprotein A2. Mol Biol Cell 21: 3578-3589. [Crossref]
- Sun Y, Wu YX, Zhang P, Peng G, Yu SY (2017) Anti-rheumatic drug iguratimod protects against cancer-induced bone pain and bone destruction in a rat model. Oncol Lett 13: 4849-4856. [Crossref]
- Wu YX, Sun Y, Ye YP, Zhang P, Guo JC, et al. (2017) Iguratimod prevents ovariectomy‑induced bone loss and suppresses osteoclastogenesis via inhibition of peroxisome proliferator‑activated receptor‑γ. Mol Med Rep 16: 8200-8208. [Crossref]
- Kunisada T, Miyazaki M, Mihara K, Gao C, Kawai A, et al. (1998) A new human chondrosarcoma cell line (OUMS-27) that maintains chondrocytic differentiation. Int J Cancer 77: 854-859. [Crossref]
- Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, et al. (2003) Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res 63: 7679-7688. [Crossref]
- Fischer G, Aumüller T (2003) Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol 148: 105-150. [Crossref]
- Dongsheng Z, Zhiguang F, Junfeng J, Zifan L, Li W (2010) Cyclophilin A Aggravates Collagen-Induced Arthritis via Promoting Classically Activated Macrophages. Inflammation 40: 1761-1772. [Crossref]
- Wang L, Wang CH, Jia JF, Ma XK, Li Y, et al. (2010) Contribution of cyclophilin A to the regulation of inflammatory processes in rheumatoid arthritis. J Clin Immunol 30: 24-33. [Crossref]
- Wang L, Jia J, Wang C, Ma X, Liao C, et al. (2013) Inhibition of synovitis and joint destruction by a new single domain antibody specific for cyclophilin A in two different mouse models of rheumatoid arthritis. Arthritis Res Ther 15: R208. [Crossref]
- Guo M, Shen J, Kwak JH, Choi B, Lee M, et al. (2015) Novel role for cyclophilin A in regulation of chondrogenic commitment and endochondral ossification. Mol Cell Biol 35: 2119-2130. [Crossref]
Editorial Information
Editor-in-Chief
Ivan Gout
University College London
Ricardo H. Alvarez
Cancer Treatment Centers of America
Article Type
Review Article
Publication history
Received date: November 20, 2017
Accepted date: December 11, 2017
Published date: December 14, 2017
Copyright
© 2017 Ooka S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
Ooka S, Kurokawa MS, Yokoyama M, Arito M, Sato T(2017) Effects of iguratimod on protein profiles of chondrosarcoma cells. Integr Mol Med 4: DOI: 10.15761/IMM.315

Figure 1. Protein profiles of OUMS-27 cells treated with IGU. Representative results of 2D-DIGE of proteins from OUMS-27 cells cultured in the presence (left upper) or absence (right upper) of 100 μM IGU for 24 hours are shown. Similarly, those cultured in the presence (left lower) or absence (right lower) of 100 μM IGU for 6 days are shown.

Figure 2. Time course of each protein expression of OUMS-27 cells in the presence of IGU. A, B. Nine out of the 11 protein spots and 4 out of the 5 protein spots, intensity of which were increased to 1.3-fold or more (A) and decreased to -1.3-fold or less (B) in the 24 hour-treatment, respectively (Table 1), were detected both in the 24 hour- and 6 day-treatments and applied for time course analysis. C, D. 14 out of the 22 protein spots and 11 out of the 15 protein spots, intensity of which were increased to 1.3-fold or more (C) and decreased to -1.3-fold or less (D) in the 6 day-treatment, respectively (Table 1), were detected both in the 24 hour- and 6 day-treatments and applied for time course analysis. Control samples (Cont) were proteins from OUMS-27 cells cultured without IGU for 24 hours (A, B) and 6 days (C, D).

Figure 3. Locations of the identified protein spots on a 2D-DIGE gel. Proteins included in 15 spots out of the 37 spots that showed ±1.3-fold or more change in intensity by the 6 day-treatment with IGU were identified. A representative result of the standard sample is shown.

Figure 4. Comparisons of spot intensity of the identified proteins between IGU-treated and non-treated conditions. Representative results of the identified 15 protein spots and intensity difference of those spots between the IGU-treated and non-treated (control) cells are shown. Error bars, standard deviations, *p<0.05.

Figure 5. Interaction of the identified proteins. Result of a pathway analysis. Symbols in red and green show the identified proteins intensity of which were increased and decreased by the IGU-treatment, respectively. Solid lines and dotted lines show direct and indirect interactions, respectively. Arrows indicate activation.
Table 1. Change of the OUMS-27 protein profile by the treatment with IGU
Culture periods |
Total number of detected
protein spots |
Number of protein spots
with different intensity
(p<0.05) |
Fold difference, IGU/Control |
Number of the
protein spots |
24 hours |
776 |
41 |
x ≥ 1.5 1.5 > x ≥ 1.3 1.3 > x ≥ -1.3 -1.3 > x ≥ -1.5 -1.5 ≥ x |
2 9 25 3 2 |
6 days |
803 |
150 |
x ≥ 1.5 1.5 > x ≥ 1.3 1.3 > x ≥ -1.3 -1.3 > x ≥ -1.5 -1.5 ≥ x |
7 15 113 11 4 |
Table 2. Identification of protein spots, intensity of which was changed by the treatment with IGU
Spot
ID |
Difference (folds) |
Proteins |
Accession ID |
MW (kDa) |
pI |
Mascot
scores |
Coverage (%) |
Confirmed sequences (Mascot ion scores) |
Theoret |
Theoret |
Observed |
Observed |
|
|
|
1072 |
1.67 |
NACHT, LRR and PYD domains-containing protein 3 |
NLRP3_HUMAN |
118.1 |
23 |
6.22 |
6.1 |
71 |
13 |
661MDHMVSSFCIENCHR675 (11) |
487 |
1.53 |
Cyclin-L1 (CCNL1) |
CCNL1_HUMAN |
59.6 |
60 |
10.7 |
6.2 |
75 |
18 |
99LPQVAM*ATGQVLFHR113 (21) |
1033 |
1.50 |
Transmembrane protein 95 (TMM95) |
TMM95_HUMAN |
19.6 |
24 |
8.98 |
7.8 |
57 |
37 |
31LARLCSQMEAR41 (2)
79EAVSSLPSYWSWLRK93 (3) |
763 |
1.40 |
Transcription factor HIVEP2
(HIVEP2) |
ZEP2_HUMAN |
228.9 |
41 |
6.5 |
5.8 |
65 |
8 |
436NALSVTTTSQERAAM*GR452 (11) |
445 |
1.37 |
Tuftelin-interacting protein 11
(TFIP11) |
TFP11_HUMAN |
96.8 |
64 |
5.45 |
5.8 |
61 |
12 |
262ELSQVKVIDMTGR274 (25)
387M*QPDCSNPLTLDECAR402 (7) |
924 |
1.34 |
14-3-3 protein epsilon (YWHAE) |
1433E_HUMAN |
29.2 |
29 |
4.63 |
4.5 |
189 |
18 |
131YLAEFATGNDR141 (54)
131YLAEFATGNDRK142 (16)
154AASDIAMTELPPTHPIR170 (77)
216DSTLIMQLLR225 (18) |
1107 |
1.34 |
Cyclophilin A (PPIA) |
PPIA_HUMAN |
18.0 |
21 |
7.68 |
8.0 |
74 |
10 |
2VNPTVFFDIAVDGEPLGR19 (66) |
87 |
-1.35 |
Hypoxia up-regulated
protein 1 (HYOU1) |
HYOU1_HUMAN |
111.3 |
98 |
5.16 |
5.3 |
66 |
12 |
439DAVVYPILVEFTR451 (24) |
599 |
-1.34 |
ATP synthase subunit alpha,
mitochondrial
(ATP5A1) |
ATPA_HUMAN |
59.7 |
53 |
9.16 |
8.0 |
185 |
11 |
134TGAIVDVPVGEELLGR139 (72)
335EAYPGDVFYLHSR347 (72)
403GIRPAINVGLSVSR416 (15) |
376 |
-1.49 |
Keratin, type II cytoskeletal 1, (KRT1)
Keratin, type II cytoskeletal 6B (K2C6B) |
K2C1_HUMAN
K2C6B_HUMAN |
66.0 66.0 |
69 |
8.15 8.09 |
6.9 |
106 82 |
9 1 |
377YEELQITAGR386 (78)
360YEELQITAGR369 (78) |
784 |
-1.31 |
Heterogenous nuclear ribonucleoproteins A2/B1
(hnRNP A2/B1) |
ROA2_HUMAN |
37.4 |
39 |
8.97 |
9.5 |
99 |
8 |
23LFIGGLSFETTEESLR38 (38)
214GGGGNFGPGPGSNFR228 (46) |
797 |
-1.35 |
Heterogenous nuclear ribonucleoproteins A2/B1
(hnRNP A2/B1) |
ROA2_HUMAN |
37.4 |
38 |
8.97 |
8.8 |
256 |
23 |
23LFIGGLSFETTEESLR38 (52)
154GFGFVTFDDHDPVDK168 (83)
214GGGGNFGPGPGSNFR228 (64) |
794 |
-1.42 |
Heterogenous nuclear ribonucleoproteins A2/B1 (hnRNP A2/B1)
|
ROA2_HUMAN |
37.4 |
38 |
8.97 |
8.7 |
129 |
13 |
23LFIGGLSFETTEESLR38 (47)
214GGGGNFGPGPGSNFR228 (60) |
773 |
-1.52 |
Heterogenous nuclear ribonucleoproteins A1
(hnRNP A1)
Heterogenous nuclear ribonucleoproteins A1-like 2
(hnRNP A1L2) |
ROA1_HUMAN RA1L2_HUMAN |
38.7 34.2 |
40 |
9.17 9.08 |
9.2 |
77 61 |
24
23 |
16LFIGGLSFETTDESLR31 (9)
337SSGPYGGGGQYFAKPR352 (5)
16LFIGGLSFETTDESLR31 (9) |
775 |
-1.60 |
Heterogenous nuclear ribonucleoproteins A1(hnRNP A1)
|
ROA1_HUMAN |
38.7 |
39 |
9.17 |
8.8 |
62 |
8 |
15KLFIGGLSFETTDESLR31(15)
16LFIGGLSFETTDESLR31 (25) |
37 protein spots, intensity of which changed to ±1.3-fold or more by the 6 day-treatment with IGU, were selected for the protein identification. Proteins of 15 out of the 37 spots were identified. MW, molecular weights.
*Oxidation of methionine.





