Abstract
Glioblastomas (GBMs) are fatal WHO grade IV astrocytomas with a median survival of 14.6 months. Immunohistochemical (IHC) studies indicate that cyclophilin A (CYPA), and its receptor CD147 are elevated in GBM, but the impacts on disease progression are poorly understood. We determined their occurrence in diffuse astrocytomas (WHO grades II-IV) using IHC and evaluated how gene expression affects patient outcomes in The Cancer Genome Atlas (TCGA) and REMBRANDT datasets (GBM and Low-Grade-Glioma: LGG). While IHC staining of CYPA was evident across all tumour grades, CD147 staining increased with tumour grade. However, while analysis of the TCGA grade II/III tumour dataset showed a trend towards high CD147 gene expression and reduced patient survival (HR=2.34, p=0.064, n=132), the results were not statistically significant. By contrast, analysis of the TCGA GBM dataset showed that high CYPA expression was significantly associated with reduced survival (HR=1.30, p=0.009; n=593). Furthermore, Kaplan-Meier analysis of both the TCGA and REMBRANDT (p=0.052; n=176) GBM datasets showed a comparable reduction in survival of about 3-months for patients with high CYPA expression. We found that CYPA gene amplification (≥2.2 copies) occurred in 72% of patients (REMBRANDT GBM dataset) and was correlated with reduced survival (log-rank p = 0.022). Finally, we observed a correlation between CYPA and CD147 gene expression in grade II/III (R=0.481, p=1.81x10-6) and grade IV (R=0.132; p=0.0012) tumours. Together, our data identify CYPA and CD147 as potential pathogenic determinants in glioblastoma.
Key words
glioblastoma, astrocytoma, cyclophilin A, CD147, gene expression, immunohistochemistry, TCGA, REMBRANDT
Introduction
Glioblastomas (GBMs), otherwise known as grade IV (WHO) astrocytomas, are highly proliferative and invasive primary brain cancers. Despite aggressive surgery, radiotherapy and adjuvant chemotherapy (Temozolomide, TMZ), tumour re-occurrence is often rapid, leaving current median survival at just 14.6 months [1]. The lack of progress in treating GBMs has meant that patients face disproportionate levels of morbidity and mortality compared to other cancers [1]. Arguably, improving survival rates in GBM patients will be contingent on finding clinically relevant disease determinants for therapeutic targeting. Cyclophilin A (CYPA) and its receptor CD147, which we previously reported are potent and interacting cytoprotectants [2-5], are two cancer determinants, which were recently linked to GBM [6-14].
Cyclophilin A, an ubiquitous intracellular/extracellular isomerase and cyclosporine (CsA) target, is implicated in protein folding/trafficking, cell-signalling, apoptosis and cytoprotection [2,3,15]. Inhibiting CYPA blocks IL-8 release and tumour growth of U87 cells [10], while enhancing cisplatin toxicity in C6 glioma cells [9,16]. Further, the expression of its receptor CD147, a glycosylated membrane-bound immunoglobulin [3], is reportedly elevated in 80% of paediatric and adult gliomas [8,11]. CD147 expression reportedly increases with tumour grade [6,17], and in paediatric gliomas, correlates with matrix metalloproteinase 2 (MMP2) and CD44 expression [8,11]. Finally, in vitro studies have revealed that CD147 knockdown inhibits glioma cell migration and MMP secretion [18]. Importantly, these CD147 related functions are likely to influence the invasive properties of GBM cells. While recent studies report that CYPA and CD147 are elevated in GBM [10,13,14], their role in disease progression and occurrence in lower grade gliomas remains unclear. Added to this, evidence has emerged indicating that CYPA and CD147 can interact to promote tumourigenesis [19].
To date, the limited numbers of studies reporting CYPA and CD147 expression in GBM have predominantly relied on immunohistochemical (IHC) data. While useful, IHC is only semi-quantitative, with staining levels confined to simple categories (e.g. 0-3). Thus, adopting IHC to correlate candidate tumour gene expression and patient survival comes with caveats. Also, while increased CD147 staining is reportedly prognostic [14] in GBM, staining intensity may not necessarily equate with actual changes in protein levels. This is because the contribution of CD147 to tumorigenesis appears to correlate with the degree of receptor glycosylation [17,20,21], which is independent of protein expression. Similarly, while the cytokine behaviour of CYPA may be a contributing factor in GBM pathogenesis, as is reported for other malignancies [12,19,22], its extracellular location means it is unlikely to be detected by IHC.
The availability of high quality tumour specific genetic data in large depositories like The Cancer Genome Atlas (TCGA) and REMBRANDT can provide powerful quantitative evidence to corroborate and/or inform the findings of IHC studies. Thus, this study evaluated CYPA and CD147 expression in GBM and in low grade astrocytoma using both strategies.
Materials and methods
Patient tissue
The Western Australian, state government pathology service, Path West, provided archived astrocytoma (confirmed pathology) and non-neoplastic brain tissue. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the declaration of Helsinki and the permission to use human tissue was granted by the Sir Charles Gairdner Group, Human Research Ethics Committee (approval #2009-016).
Immunohistochemistry
Gliomas were classified and graded according to current World Health Organisation (WHO) Guidelines [23]. Specimens were received fixed in 10% buffered neutral formalin, assessed macroscopically and routinely processed into paraffin embedded blocks. Following processing, 4μm sections of tissue were cut, transferred onto glass slides, deparaffinized, re-hydrated and subjected to antigen retrieval. Sections were probed with rabbit polyclonal anti-CYPA antibody (1:3000, Biomol, USA) or mouse monoclonal anti-CD147 (1:10000, Santa Cruz, USA). Anti-mouse and anti-rabbit HRP-tagged secondary antibodies were used according to the manufacturers instructions (EnVisionTM DAKO, USA). Sections were stained with 3,3’-diamino-benzidine and counterstained with Haematoxylin.
Analysis of tissue immunoreactivity
Tissue sections were scored according to the method of Gu et al [8]. In brief, the numbers of cells showing membrane and cytoplasmic staining from ten representative microscope fields were totalled. The percentage immunoreactivity was determined for each tissue section and a semi-quantitative value was derived according to the following scheme: 0 = absent/<5%; 1+ = weak/6-25%; 2+ = moderate/26-50% and; 3+ = strong/>51%.
Publicly available datasets
To investigate the clinical significance of CYPA and CD147 mRNA overexpression in GBM, we downloaded the clinicopathologic and intratumoral mRNA expression data from 593 Glioblastoma (GBM) and 132 Low Grade Glioma (LGG) astrocytoma patient datasets at The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp; accessed October 2013). The intratumoral mRNA levels provided were measured using Illumina HiSeq 2000 RNA Sequencing Version 2 and were subsequently normalised. These datasets were previously validated [24,25] and certified for use without limitation (http://cancergenome.nih.gov/publications/publicationguidelines). The GBM cohort of 593 patients comprised 360 men and 233 women (mean age 57.7 years), with 457-recorded deaths from all causes, at the time of the study. The cohort of 132 patients with astrocytoma consisted of 70 men and 62 women (mean age 44.5 years) comprising grade II (n=43) and grade III (n=89) tumours, with 26-recorded deaths from all causes, at the time of the study. It should be noted that the TCGA datasets denoted as low-grade or lower grade glioma (LGG) included WHO grade II/III astrocytomas. Data from the Repository for Molecular BRAin Neoplasia DaTa (REMBRANDT) National Cancer Institute 2005, was analysed using the REMBRANDT web-based interface for simple calculations, accessed 2014 April 30. The original REMBRANDT database is now accessible via the link: https://gdoc.georgetown.edu/gdoc/workflows/index.
Statistical and bioinformatics analyses
The GBM and astrocytoma cohorts from The Cancer Genome Atlas (TCGA) [26] were statistically analysed using the R programming environment [27]. Kaplan-Meier survival curves showing the prognostic significance of CYPA and CD147 overexpression, were produced using the R “survival” package. Cox regressions were calculated using the R “coxph” function from the “survival” package with ties addressed by Efron’s method. For both Cox regressions and Kaplan-Meier survival curves, all deaths were used as an endpoint. As indicated, analyses were adjusted for age and gender. The validity of proportional hazards assumptions were assessed by scaled Schoenfeld residuals [21]. Correlation coefficients were calculated using the R “cor.test” function and a simple linear model fitted to CYPA and CD147 expression data using the “lm” function.
Kaplan-Meier and gene copy number data from REMBRANDT were analysed using the REMBRANDT web-based interface and downloaded without manipulation (Figure 3b; Figure 4). Copy number was measured using the SNP 100K chip data. The REMBRANDT GBM data set provided expression intensities for all 8 CYPA probe tested. The median expression intensity for all CYPA Affymetrix probes was 8.11 (range: 6.7 - 9.7; standard deviation of 1.3), meaning all probes reported a similar CYPA expression. The CYPA gene copy number versus patient survival data reported in Figure 4, was derived from probe 217602_at, which had an expression intensity of 8.97, was close to the mean intensity value for all probes.
Microscopy and imaging
Microscopy and imaging: Image acquisition was performed using an Olympus IX70 microscope fitted with a cooled CCD digital camera (DP70; Olympus, Japan) under software control (DP controller; Olympus).
Results
Staining of CYPA and CD147 in astrocytoma (WHO grades II-IV)
Astrocytoma tumour sections (N=30) were stained for CYPA and CD147 (Table 1, Figure 1). CD147 staining was moderate-to-strong in GBM tissue, moderate in WHO grade III analplastic astrocytoma and mostly undetectable in WHO grade II diffuse astrocytomas (N=13). Based on these data, CD147 staining increased with tumour grading. Cyclophilin A staining was moderate-to-strong in GBM (N=13), moderate in grade III (N=3) and weak-to-moderate in grade II (N=13) astrocytoma. Compared to CD147, while CYPA staining increased with tumour grade, the delineation between grades was less marked.
Table 1. Summary of CYPA and CD147 staining of 30 astrocytoma tissues (WHO grades II-IV).
Tumour Grade |
Patients (N) |
CYPA |
CD147 |
II |
13 |
Weak (7);
Moderate (4);
Strong (2) |
Absent (12); Moderate (1) |
III |
3 |
Moderate (2);
Strong (1) |
Moderate (3) |
IV (GBM) |
14 |
Weak (2);
Moderate (9);
Strong (3) |
Absent (3);
Weak (3);
Moderate (3);
Strong (5) |
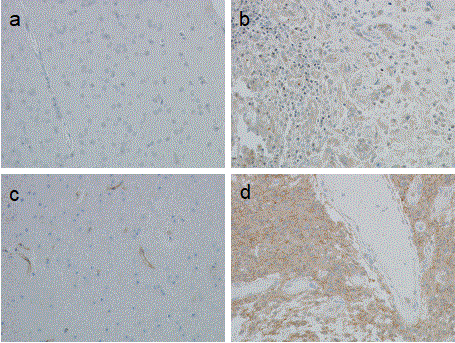
Figure 1. Immunohistochemical staining of glioblastoma tissue. Formalin fixed, paraffin embedded sections (4 μm) were probed with either anti-CYPA or anti-CD147 antibodies and stained with DAB and counterstained with haematoxylin (image magnification 100x). Panels a) Non-neoplastic brain tissue (anti-CYPA 1:3000); b) GBM tissue (anti-CYPA 1:3000); c) Non-neoplastic brain tissue (anti-CD147, 1:10000), note the endothelial cell staining; d) GBM tissue (anti-CD147, 1:10000).
Analysis of CYPA and CD147 gene expression in grade II/III astrocytomas
To test our IHC findings, we analysed grade II/III astrocytoma (N=132) expression data (low-grade glioma: LGG cohort, accessed April 2014) from the The Cancer Genome Atlas (TCGA) network [28]. For both genes, there was a slight (but non-significant), 1.04 fold higher expression level in grade III tumours compared to grade II tumours (CYPA, p=0.25; CD147, p=0.498; n=132). Cox-regression analysis of CYPA expression (adjusted for age and gender) showed that the slight difference between grade II and grade III tumours did not affect overall survival (HR=0.9, p=0.81; n=132, Table 2).
Table 2: Multivariate Cox regression analyses showing overall survival by gene expression according to tumour grade.
Variable |
n
patients |
n
deaths |
HR (adjusted for age and gender) |
p-value |
95% confidence interval of HR |
|
Grade 2 & 3 |
132 |
26 |
|
|
|
|
High CD147 |
|
|
2.34 |
0.064 |
0.95-5.75 |
|
High CYPA |
|
|
0.90 |
0.81 |
0.40-2.03 |
|
|
|
|
|
|
|
|
Grade 2 |
43 |
2 |
Too few |
|
|
|
High CD147 |
|
|
deaths |
|
|
|
High CYPA |
|
|
|
|
|
|
|
|
|
|
|
|
|
Grade 3 |
89 |
24 |
|
|
|
|
High CD147 |
|
|
2.03 |
0.16 |
0.76-5.45 |
|
High CYPA |
|
|
0.97 |
0.94 |
0.41-2.31 |
|
|
|
|
|
|
|
|
By contrast, CD147 gene expression in grade III astrocytoma correlated with reduced patient survival (HR 2.34, p=0.064; n=132) when compared to grade II astrocytoma. Within the astrocytoma grade III cohort, Kaplan-Meier analysis of patient survival returned a hazard ratio of 1.92 (p=0.14, n=89) for high expression of CD147 compared to low CD147 gene expression (Figure 2). In this instance, the number of patients was relatively small and the results were not significant.
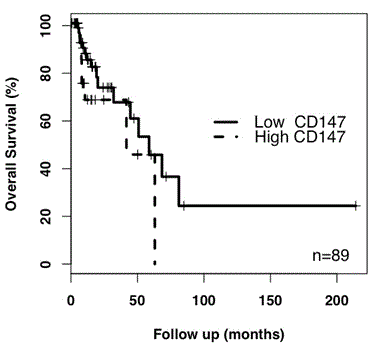
Figure 2. Kaplan-Meier analysis of CD147 expression in astrocytoma grade III. High CD147 expression returned a HR value of 1.92 (p=0.14, n=89) relative to low CD147 expression. As only 2 deaths were recorded for grade II astrocytoma patients, a Kaplan-meier analysis was not performed.
A Pearson's correlation coefficient determination revealed a strong correlation between CD147 and CYPA expression in astrocytoma grade III (R = 0.481 and a p-value of 1.81 x 10-6; data not shown). In other words, high CD147 gene expression was associated with high CYPA gene expression.
Analysis of CYPA and CD147 gene expression in GBM
Cox-regression analysis of CYPA and CD147 gene expression data from the TCGA GBM cohort (N=593; Table 3) using multivariate analysis (adjusted for age and gender) showed that high CYPA expression was associated with reduced patient survival compared to low CYPA expression (Hazard Ratio: HR, of 1.30, p=0.009, N=593).
Table 3: Univariate and multivariate Cox regression analysis of overall survival by gene expression in patients with glioblastoma.
Gene expression
(High vs Low) |
HRa (unadjusted) |
95% confidence interval of HR (unadjusted |
HRb
(adjusted for age and gender) |
95% confidence interval of HR (adjusted) |
CD147 |
1.06 (p=0.661) |
0.82-1.37 |
1.06 (p=0.860) |
0.76-1.26 |
CYPA |
1.24 (p=0.031) |
1.02-1.51 |
1.30 (p=0.009) |
1.07-1.58 |
Kaplan-Meier survival plots for CYPA showed that high expression decreased median patient survival by approximately 3 months (Figure 3a). To test these findings, we analysed CYPA and CD147 gene expression (N=176) in a second GBM dataset, the REMBRANDT database.
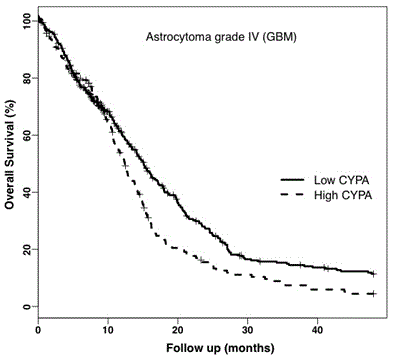
Figure 3a. Kaplan-Meier Survival Plot of CYPA Gene Expression in GBM. Analysis of the TCGA GBM dataset showed CYPA gene expression correlated with decreased overall percent survival (log-rank p = 0.009) adjusted for age and gender. The solid and dotted plot-lines correspond to low (399 patients) and high (194 patients) CYPA gene expression respectively.
We found strong agreement with the TCGA dataset, in that higher CYPA expression was associated with a reduction in median patient survival (p=0.052) of 85 days or 2.75 months (Figure 3b). CD147 gene expression was not associated with reduced patient survival (HR 1.06 p=0.860, 0.76-1.26).
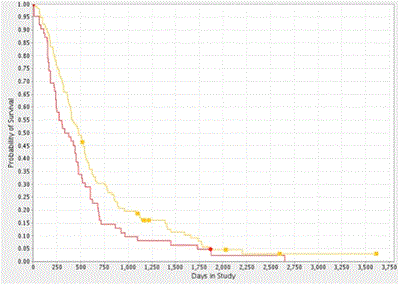
Figure 3b. Kaplan-Meier Survival Plot of CYPA Gene Expression in GBM. Analysis of the REMBRANDT GBM dataset showed that high CYPA gene expression was correlated with decreased probability of survival (log-rank p = 0.052). The yellow and red plot-lines correspond to intermediate (114 patients) and upregulated (62 patients) CYPA gene expression respectively.
CYPA and CD147 gene expression were highly correlated (Pearson’s correlation score was 0.132; p=0.002, n=593; Figure 3c).
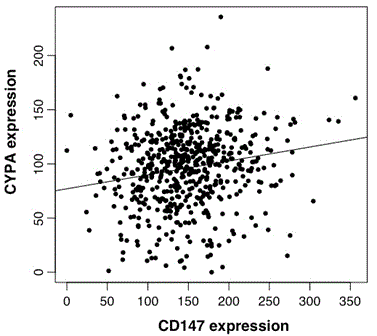
Figure 3c. Scatter-plot of CYPA and CD147 expression in GBM. Analysis of CYPA and CD147 TCGA data n=593, cor=0.129, p=0.002.
CYPA gene copy number correlates with patient survival
The consistent distribution pattern of CYPA gene expression, viz. 33% and 67% (high versus low, respectively) and patient survival observed in both GBM datasets suggested a potential genetic origin. To explore this possibility, we derived a Kaplan-Meier survival plot as function of CYPA gene copy number using data from the REMBRANDT dataset. We found that the CYPA gene was amplified by ³ 2.2 copies in 72.2% of patients (determined using the AFFymetrix reporter: 217602_at) and that this increase significantly correlated with reduced patient survival (log rank p-value of 0.022, n=90; Figure 4). It should be noted that the AFFymetrix reporter: 217602_at maps to the first intronic sequence of the human PPIA gene.
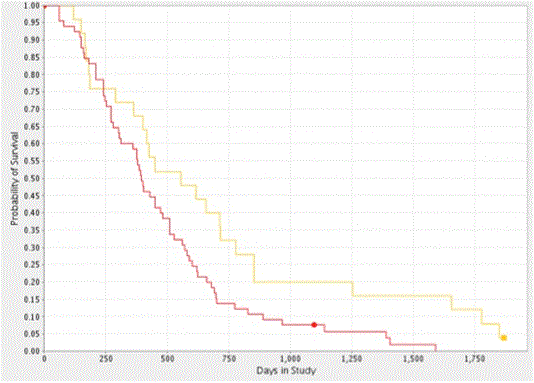
Figure 4. Kaplan-Meier Survival Plot of CYPA gene Copy Number in GBM. Analysis of the REMBRANDT GBM dataset showed CYPA gene copy amplification (>2.2 copies) decreased the probability of patient survival (log-rank p = 0.022). The yellow and red plot-lines correspond to intermediate (25 patients) and upregulated (65 patients) samples.
Discussion
A key finding of this study is that CYPA gene expression is predictive of patient outcome. Specifically, multivariate cox-regression and Kaplan-Meier analysis of CYPA gene expression in the TCGA and REMBRANDT GBM datasets revealed a statistically significant relationship between high CYPA expression and reduced overall patient survival. We found substantial consistency across both GBM datasets, where a third of all patients had tumours with high CYPA expression and that median survival among this group was reduced by about 3 months. By contrast, a comparison of CYPA staining in GBM and low-grade astrocytoma was less clear, although a weak correlation was suggested. For example, while staining intensity was generally robust in GBM and grade III astrocytomas, CYPA staining was also evident in grade II astocytomas. Similarly, CYPA gene expression showed no association between tumour grade and overall patient survival in the TCGA LGG dataset. Thus, while differential CYPA gene expression can predict GBM patient outcome, it cannot distinguish between low-grade glioma (grade II/III astrocytoma) and GBM. While elevated CYPA expression has been previously noted in GBM tissue using IHC [10], this is the first study to link especially high gene expression with reduced survival.
As CYPA is encoded on chromosome 7, a chromosome frequently amplified in GBM isolates (29), we asked if CYPA amplification could account for the difference in gene expression observed. However, we found that increased CYPA copy number, which occurred in >72% of patients (according to REMBRANDT), did not equate with the 33% of patients that showed particularly high CYPA gene expression. Not surprisingly, the genetic origins of CYPA gene expression in GBM are more complex and are likely to involve regulatory, transcriptional and gene dosage effects.
Consistent with previous reports [13,14,30] we observed robust CD147 staining in GBM tissue. However, we found no difference in patient survival (TCGA and REMBRANDT) between high and low CD147 gene expression. Our findings are in line with a colorectal cancer study that showed an association between CD147 staining and tumour severity, but not between mRNA levels and tumour severity [31]. The extent to which CD147 is glycosylated, which purportedly increases with tumour grade and virulence [21], may account for the disparity between our gene expression data and the findings described in IHC studies [13,14,30]. In this instance, the correlation between differential staining intensity and malignancy may arise if the antibodies used exhibited a greater affinity for glycosylated CD147. This notion is supported by the work of Reithdorf et al (2006) who showed that removing specific carbohydrate moieties from glycosylated CD147 could alter and even abolish antibody reactivity [17]. Nonetheless, as CD147 staining was evident in grade III astrocytomas and GBMs, but virtually absent in grade II astrocytomas, we suggest that CD147 staining could be used to distinguish grade II astrocytomas from grade III astrocytomas and GBMs. This view is also supported by our multivariate analysis of CD147 gene expression of the LGG (TCGA) dataset which showed that CD147 expression was associated with a HR of 2.34 for grade III tumours (p=0.064; n=132) compared to grade II tumours. Within the astrocytoma grade III cohort, Kaplan-Meier analysis of patient survival returned a hazard ratio of 1.92 (p=0.14; n=89) for high expression compared to low CD147 expression. However, the number of patients was relatively small and the value was not significant.
This study also revealed a correlation between CD147 and CYPA expression in both the GBM (R=0.132; p=0.002) and LGG datasets (R = 0.481; p=1.81 x 10-6). To our knowledge this study is the first study to report such a finding in GBM, or for any human disease, including other malignancies. This is despite considerable experimental evidence that CYPA and CD147 participate and/or interact in many disease related pathways [19]. Although interesting, the association between CYPA and CD147 expression remains speculative as the molecular basis for this interaction was not investigated. Nonetheless, previous studies suggest that transcriptional activation of both CYPA and CD147, could arise via the hypoxia inducible factor (HIF) [32,33] and/or the transcription factor Sp1 [33]. Evidence also suggests that CYPA directly increases CD147 expression, at least in cholangiocarcinoma cells [19]. Finally, that CYPA and CD147 share a regulatory pathway(s) warrants further investigation.
As HIF mediates transcriptional activation of CYPA expression, a potentially novel pathway linking CYPA and CD147 could involve hypoxic and/or oxidative stress induced secretion of CYPA [34,35]. As activation of the HIF pathway is a driver of tumour growth and angiogenesis in GBM [36,37], it would be interesting to determine if GBM cells, like other cancer cells [19], can secrete CYPA. In this scenario, extracellular CYPA could contribute to GBM tumourigenesis by activating pro-survival signaling [2,4,38], cell proliferation [19], cell invasion/migration [39-41] and MMP release [42], presumably via the CD147 receptor. Interestingly, extracellular CYPA has been detected in the CSF of patients with diffuse pontine glioma [43] and in an experimental model of prion disease [44].
While the true impacts of CYPA on GBM progression awaits full investigation, studies have identified potent tumourigenic properties for CYPA that include: abrogating hypoxic stress [2,3], enhancing chemoresistance [10,45,46] supporting cell-proliferation and invasiveness [10,46,47] and invoking inflammatory/angiogenic cytokine release [48-50]. Likewise, the tumourigenic consequences of CD147 receptor activation (either by homo-dimer formation or CYPA binding) are also extensive and include the activation of the MAPK (ERK1/2, p38, and JNK), PI3/Akt and NF-κB signalling pathways which can lead to pro-survival signalling, cytokine release, cell migration and proliferation [2,3]. Added to these, CD147 can also exert non-CYPA related tumourigenic impacts such augmenting CD44/EGFR signaling and critically, in trafficking the oncogenic monocarboxylate proteins, MCT1 and MCT4 [11]. Finally, a recent study showed that CYPA actually promotes glioma-initiating cell stemness, self-renewal, proliferation and radioresistance via Wnt/B-catenin signaling [51]. If confirmed, it would demonstrate a potentially critical role for CYPA in glioblastoma development, recurrence and treatment resistance, consistent with the present findings.
Conclusion
Our findings highlight the need to consider both IHC and gene expression data when assessing the relevance of a given disease determinant/marker. Our findings also reveal that CYPA gene expression is an independent prognostic factor of patient outcome in GBM and a potential therapeutic target that warrants further investigation.
Acknowledgements
This work was supported by the Perron Institute for Neurological and Translational Science (WANRI) and PathWest. The authors declare they have no conflicting interests. The authors acknowledge the contributions to this work by the TCGA Research Network (data set) and National Cancer Institute, REMBRANDT (2005), the specimen donors and other research groups involved in the collection and generation of data. The authors also acknowledge the technical support of Michael Platten and Russell Johnsen. Finally, we would like to express our gratitude to Professor Frank Mastaglia for his insightful comments and suggestions during the preparation of this manuscript.
2021 Copyright OAT. All rights reserv
Author Contributions
KW, TJ and SB conducted experimental work and drafted the manuscript. PC conducted all TCGA and REMBRANDT analyses. VM, PC, NK and PR drafted the manuscript.
References
- Huse JT, Holland EC (2010) Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 10: 319-331. [Crossref]
- Boulos S, Meloni BP, Arthur PG, Majda B, Bojarski C, et al. (2007) Evidence that intracellular cyclophilin a and cyclophilin a/cd147 receptor-mediated erk1/2 signalling can protect neurons against in vitro oxidative and ischemic injury. Neurobiology of disease 25: 54-64. [Crossref]
- Kanyenda LJ, Verdile G, Boulos S, Krishnaswamy S, Taddei K, et al. (2011) The dynamics of CD147 in Alzheimer's disease development and pathology. J Alzheimers Dis 26: 593-605. [Crossref]
- Kanyenda LJ, Verdile G, Martins R, Meloni BP, Chieng J, et al. (2013) Is cholesterol and amyloid-beta stress induced cd147 expression a protective response? Evidence that extracellular cyclophilin a mediated neuroprotection is reliant on cd147. J Alzheimers Dis 39:545-56 [Crossref]
- Meloni BP, Tilbrook PA, Boulos S, Arthur PG, Knuckey NW (2006) Erythropoietin preconditioning in neuronal cultures: Signaling, protection from in vitro ischemia, and proteomic analysis. J Neurosci Res 83: 584-593. [Crossref]
- Sameshima T, Nabeshima K, Toole BP, Yokogami K, Okada Y, et al. (2000) Expression of emmprin (cd147), a cell surface inducer of matrix metalloproteinases, in normal human brain and gliomas. International journal of cancer. Int J Cancer 88: 21-27. [Crossref]
- Li H, Che J, He M, Hui XH, Cai BW, et al. (2007) Expression of cd147 and mmp-2 in human gliomas and its correlations with prognosis. Sichuan Da Xue Xue Bao Yi Xue Ban 38: 396-399. [Crossref]
- Gu J, Zhang C, Chen, R, Pan J, Wang Y, et al. (2009) Clinical implications and prognostic value of emmprin/cd147 and mmp2 expression in pediatric gliomas. Eur J Pediatr 168: 705-710. [Crossref]
- Han X, Yoon SH, Ding Y, Choi TG, Choi WJ, et al. (2010) Cyclosporin A and sanglifehrin A enhance chemotherapeutic effect of cisplatin in C6 glioma cells. Oncol Rep 23: 1053-1062. [Crossref]
- Sun S, Wang Q, Giang A, Cheng C, Soo C, et al. (2011) Knockdown of cypa inhibits interleukin-8 (il-8) and il-8-mediated proliferation and tumor growth of glioblastoma cells through down-regulated nf-kappab. J Neurooncol 101: 1-14. [Crossref]
- Maria B, Gupta N, Gilg AG, Abdel W, Leonard AP, et al. Targeting hyaluronan interactions in spinal cord astrocytomas and diffuse pontine gliomas. J Child Neurol 23: 1214-1220. [Crossref]
- Lee J (2010) Role of cyclophilin a during oncogenesis. Arch Pharm Res 33: 181-187. [Crossref]
- Tian L, Zhang Y, Chen Y, Cai M, Dong H, et al. (2013) Emmprin is an independent negative prognostic factor for patients with astrocytic glioma. PloS one 8: e58069. [Crossref]
- Yang M, Yuan Y, Zhang H, Yan M, Wang S, et al. (2013) Prognostic significance of CD147 in patients with glioblastoma. J Neurooncol 115: 19-26. [Crossref]
- Zhu C, Wang X, Deinum J, Huang Z, Gao J, et al. (2007) Cyclophilin a participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J Exp Med 204: 1741-1748. [Crossref]
- Choi JW, Schroeder MA, Sarkaria JN, Bram RJ (2014) Cyclophilin b supports myc and mutant p53-dependent survival of glioblastoma multiforme cells. Cancer Res 74: 484-496. [Crossref]
- Riethdorf S, Reimers N, Assmann V, Kornfeld JW, Terracciano L, et al. (2006) High incidence of emmprin expression in human tumors. Int J Cancer 119: 1800-1810. [Crossref]
- Liang Q, Xiong H, Gao G, Xiong K, Wang X, et al. (2005) Inhibition of basigin expression in glioblastoma cell line via antisense rna reduces tumor cell invasion and angiogenesis. Cancer Biol Ther 4: 759-762. [Crossref]
- Obchoei S, Sawanyawisuth K, Wongkham C, Kasinrerk W, Yao Q, et al. (2015) Secreted cyclophilin a mediates g1/s phase transition of cholangiocarcinoma cells via cd147/erk1/2 pathway. Tumour Biol 36: 849-859. [Crossref]
- Fan J, Wang S, Yu S, He J, Zheng W (2012) N-acetylglucosaminyltransferase iva regulates metastatic potential of mouse hepatocarcinoma cells through glycosylation of cd147. Glycoconj J 29: 323-334. [Crossref]
- Bai Y, Huang W, Ma LT, Jiang JL, Chen ZN (2014) Importance of n-glycosylation on cd147 for its biological functions. Int J Mol Sci 15: 6356-6377. [Crossref]
- Zhu D, Wang Z, Zhao JJ, Calimeri T, Meng J, et al. (2015) The Cyclophilin A-CD147 complex promotes the proliferation and homing of multiple myeloma cells. Nat Med 21: 572-580. [Crossref]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97-109. [Crossref]
- Brat DJ, Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M., et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. The New England journal of medicine 2015, 372, 2481-2498. [Crossref]
- Brennan CW1, Verhaak RG, McKenna A, Campos B, Noushmehr H, et al. (2013) The somatic genomic landscape of glioblastoma. Cell 155: 462-477. [Crossref]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061-1068. [Crossref]
- Ihak R, Gentleman RR (1996) A language for data analysis and graphics. Journal of Computational and Graphical Statistics 5: 299-314.
- Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061-1068.
- Sintupisut N, Liu PL, Yeang CH (2013) An integrative characterization of recurrent molecular aberrations in glioblastoma genomes. Nucleic Acids Res 41: 8803-8821.
- Tsai WC, Chen Y, Huang LC, Lee HS, Ma HI, et al. (2013) Emmprin expression positively correlates with who grades of astrocytomas and meningiomas. J Neurooncol 114: 281-290. [Crossref]
- Zheng Hc, Wang W, Xu XY, Xia P, Yu M, et al. (2011) Up-regulated emmprin/cd147 protein expression might play a role in colorectal carcinogenesis and its subsequent progression without an alteration of its glycosylation and mrna level. J Cancer Res Clin Oncol 137: 585-596. [Crossref]
- Huang C, Sun, Z, Sun Y, Chen X, Zhu X, et al. (2012) Association of increased ligand cyclophilin a and receptor cd147 with hypoxia, angiogenesis, metastasis and prognosis of tongue squamous cell carcinoma. Histopathology 60: 793-803. [Crossref]
- Ke X, Fei F, Chen Y, Xu L, Zhang Z, et al. (2012) Hypoxia upregulates cd147 through a combined effect of hif-1alpha and sp1 to promote glycolysis and tumor progression in epithelial solid tumors. Carcinogenesis 33: 1598-1607. [Crossref]
- Jin ZG, Melaragno MG, Liao DF, Yan C, Haendeler J, et al. (2000) Cyclophilin a is a secreted growth factor induced by oxidative stress. Circulation research 87: 789-796.
- Seko Y, Fujimura T, Taka H, Mineki R, Murayama K, et al. (2004) Hypoxia followed by reoxygenation induces secretion of cyclophilin a from cultured rat cardiac myocytes. Biochemical and biophysical research communications 317: 162-168.
- Khatua S, Peterson KM, Brown KM, Lawlor C, Santi MR, et al. (2003) Overexpression of the egfr/fkbp12/hif-2alpha pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res 63: 1865-1870. [Crossref]
- Kaur B, Khwaja FW, Severson EA, Matheny SL, Brat DJ, et al. (2005) Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol 7: 134-153. [Crossref]
- Obchoei S, Weakley SM, Wongkham S, Wongkham C, Sawanyawisuth K, et al. (2011) Cyclophilin a enhances cell proliferation and tumor growth of liver fluke-associated cholangiocarcinoma. Mol cancer 10: 102. [Crossref]
- Damsker JM, Bukrinsky MI, Constant SL, Preferential chemotaxis of activated human cd4+ t cells by extracellular cyclophilin a. J Leukoc Biol 82: 613-618. [Crossref]
- Heine SJ, Olive D, Gao JL, Murphy PM, Bukrinsky MI, et al. (2011) Cyclophilin A cooperates with MIP-2 to augment neutrophil migration. J Inflamm Res 4: 93-104. [Crossref]
- Wang CH, Dai JY, Wang L, Jia JF, Zheng ZH, et al. (2011) Expression of cd147 (emmprin) on neutrophils in rheumatoid arthritis enhances chemotaxis, matrix metalloproteinase production and invasiveness of synoviocytes. J Cell Mol Med 15: 850-860. [Crossref]
- Seizer P1, Schönberger T, Schött M, Lang MR, Langer HF, et al. (2010) EMMPRIN and its ligand cyclophilin A regulate MT1-MMP, MMP-9 and M-CSF during foam cell formation. Atherosclerosis 209: 51-57. [Crossref]
- Saratsis AM, Yadavilli S, Magge S, Rood BR, Perez J, et al. (2012) Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro Oncol 14: 547-560. [Crossref]
- Tribouillard TD, Carroll JA, Moore RA, Striebel JF, Chesebro B (2012) Role of cyclophilin a from brains of prion-infected mice in stimulation of cytokine release by microglia and astroglia in vitro. J Biol Chem 287: 4628-4639. [Crossref]
- Choi KJ, Piao YJ, Lim MJ, Kim JH, Ha J, et al. (2007) Overexpressed cyclophilin a in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Res 67: 3654-3662. [Crossref]
- Li Z, Min W, Gou J (2013) Knockdown of cyclophilin a reverses paclitaxel resistance in human endometrial cancer cells via suppression of mapk kinase pathways. Cancer Chemother Pharmacol 72: 1001-1011. [Crossref]
- Yang Y, Lu N, Zhou J, Chen ZN, Zhu P (2008) Cyclophilin a up-regulates mmp-9 expression and adhesion of monocytes/macrophages via cd147 signalling pathway in rheumatoid arthritis. Rheumatology 47: 1299-1310.
- Christofferson DE, Yuan J (2010) Cyclophilin A release as a biomarker of necrotic cell death. Cell Death Differ 17: 1942-1943. [Crossref]
- Yurchenko V1, Zybarth G, O'Connor M, Dai WW, Franchin G, et al. (2002) Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J Biol Chem 277: 22959-22965. [Crossref]
- Yurchenko V, Constant S, Eisenmesser E, Bukrinsky M (2010) Cyclophilin-cd147 interactions: A new target for anti-inflammatory therapeutics. Clin Exp Immunol 160: 305-317. [Crossref]
- Wang G, Shen J, Sun J, Jiang Z. Fan J, et al (2017) Cyclophilin A Maintains Glioma-Initiating Cell Stemness by Regulating Wnt/ß-Catenin Signaling. Clin Cancer Res 23: 6640-6649.






