Backgroundː Prognosis for patients with original glioblastoma diagnosis remains poor. Antiangiogenic therapy with Bevacizumab in first line treatment did not lead to improvement in overall survival, while progression-free survival was prolonged by 3 to 4 months, in recent phase III studies. Bevacizumab therapy in glioblastomas is not successfully associated with any prognostic biological markers. Our study investigates the correlation between the use of bevacizumab and several clinical and molecular markers measured in gliomas everyday clinical practice.
Methodsː We analyzed retrospectively 47 patients with high grade gliomas treated with bevacizumab in our medical oncology department. We examined the prognostic biomarkers used in clinical practice like IDH1 and IDH2 mutation status, EGFRvIII expression, MGMT promoter methylation status and BRAF V600E mutation status. We evaluated general patient characteristics, chemotherapy and overall survival.
Resultsː Our analysis revealed a trend towards improved overall survival in glioblastoma patients with poor prognosis and the use of bevacizumab. Overall survival in original glioblastoma diagnosis patients whose tumors carried unfavorable prognostic factors, such as EGFRvIII and unmethylated MGMT promoter, was similar to the survival of those with favorable prognostic factors. However, we were unable to identify a biomarker that was statistically associated with longer survival on bevacizumab.
Conclusionsː Our study reports some hypothesis generating hints that bevacizumab is a treatment that seems to work better in patients whose tumors carry poor prognostic factors.
glioblastoma, bevacizumab, overall survival, poor prognostic factors, biomarkers
Glioblastoma is the most common primary brain tumor in adults. Maximal safe resection is the standard of care followed by radiotherapy and concurrent temozolomide. Prognosis for patients with original glioblastoma diagnosis still remains poor, with a median survival of one year while very few second line treatment options exist [1].
Bevacizumab is a monoclonal antibody with antiangiogenic function. It binds to VEGF-A ligand and therefore affects angiogenesis by inhibiting VEGF-endothelial cell interaction. Bevacizumab is active and approved for use in many tumor types [2].
Glioblastomas are highly vascularised tumors that overexpress VEGF-A. Thus, antiangiogenic therapy was thought to be a promising choice in glioblastomas [3]. Early studies showed encouraging results [4,5], but when 2 recent phase III studies, AVA-glio and Radiation Therapy Oncology Group 0825 examined the use of bevacizumab in first line treatment of glioblastoma, overall survival was not found to be improved, while progression-free survival was prolonged by 3 to 4 months, albeit not to a statistically significant extent in RTOG 0825 [6-8]. Additionally, the use of bevacizumab in combination with lomustine in second line, though effective in the Phase II BELOB trial, did not yield a statistically significant survival benefit in the Phase III EORTC trial [9]. One possible explanation for these results may be the lack of identification of the subgroup that benefits.
Several biomarkers have been studied in gliomas [10,11]. MGMT promoter methylation status is an independent favorable prognostic factor in glioblastoma patients and an important predictive biomarker for temozolomide [12]. IDH1 and IDH2 mutations in glioma correlate with increased survival [13,14]. The epidermal growth factor receptor (EGFR) variant III is expressed in glioblastomas with aggressive behavior and resistance to chemotherapy and radiotherapy and its predictive usefulness for the use of targeted vaccines is still under evaluation [15-19]. Loss of 1p/19q chromosomes is strongly associated with oligodendroglial histology and better prognosis [20]. Astrocytic differentiation is closely related to mutations in ATRX gene that cause alternative lengthening of telomeres and subsequently create genomic instability [21,22]. Mutations in the promoter of the telomerase reverse transcriptase gene (TERT) in glioblastoma are associated with shorter overall survival [23,24]. Mutations of BRAF which is also a potential therapeutic target, seem to have an emerging role in pediatric brain tumors, but its relevance to adult glioblastoma patients still remains unknown [25,26].
Bevacizumab efficacy in glioblastomas, has not been associated with any biological markers [27,28]. In our study, we try to investigate the association of biomarkers that characterize gliomas, with the outcome on bevacizumab in high grade glioma patients.
We performed a retrospective review of patients with malignant gliomas accrued between 2006 and 2015 and collected tissue samples (tumor cell content > 75% in all cases) from 47 adult patients. All patients with original glioblastoma diagnosis received temozolomide and radiation. Bevacizumab was given to patients at 15 mg/kg q3w.
DNA & RNA was extracted from Formalin-Fixed Paraffin Embedded (FFPE) tissue using the QIAmp DNA FFPE®tissue kit (Qiagen, Germany) and High Pure FFPE RNA Micro Kit (Roche, Germany) respectively, according to the manufacturer’s instructions.
IDH1 mutation was detected either with immunohistochemistry or with PCR. Immunohistochemical staining was performed on 4μm thick formalin-fixed, paraffin-embedded (FFPE) tissue sections with a Bench Mark Ultra immunostainer (Ventana Medical Systems, Tuscon, AZ, USA). Following deparaffinization and pretreatment with Ultra Cell Conditioner I (Ventana Medical Systems, Tuscon, AZ, USA), sections were incubated with anti-IDH1R132H antibody, clone H09 (Dianova, Hamburg, Germany). For chromogenic detection, the iView DAB Detection Kit (Ventana) was used. Strong cytoplasmic staining was scored as positive.
For IDH testing with PCR, exon 4 of IDH1 gene was amplified by PCR and mutation detection was carried out by sequencing analysis. PCR conditions and primer pairs for IDH1 & IDH2 genes were previously reported [29].
Sequencing was then performed (ABI Prism 3130 sequencer) and the sample’s DNA sequence was compared with reference sequences. In all samples negative for the presence of the R132H IDH1 mutation, exon 4 of IDH2 gene was tested.
For EGFRvIII detection via Real-time PCR, cDNA synthesis followed by Real Time PCR was carried out for the detection of EGFRvIII variant using SYBR green chemistry, on a RotorGene 6000 real-time analyzer (Qiagen), as described previously [30]. Each assay was performed in triplicate. Two housekeeping genes, β-actin and ABL, were used as reference genes. In the case of positive samples, DNA sequencing analysis was performed, to confirm the specificity of the obtained Real-time PCR products.
For EGFRvIII detection via Immunohistochemistry, FFPE specimens were sectioned at 4-5 microns and mounted on positively charged slides. Slides were air-dried and then deparaffinized and hydrated through a series of xylene, graded alcohol and water stations. Slides were placed in 3% hydrogen peroxide followed by heat induced epitope retrieval in a pressure chamber while submerged in citrate buffer. Protein block was applied to the slides followed by primary antibody (EGFRvIII) incubation. Staining was visualized with DAKO EnVision Rabbit HRP and DAKO DAB. Cut off for Negative was staining for < 10% of the cells and Positive ≥ 10%. EGFRvIII Immunohistochemistry testing was performed by Clarient Diagnostic Services, Inc. and funded by Celldex Therapeutics, Inc.
The methylation pattern in the CpG island of MGMT was determined by chemical modification of unmethylated, but not methylated, cytosine to uracil, using the EpiTect Bisulfite Kit (Qiagen, Germany). A total of 10μl of bisulfite-treated DNA was carried on for PCR using specific primers for the modified methylated and the unmethylated DNA. PCR assays and primer pairs for MGMT gene were previously described [31].
BRAF exon 15 V600E mutation status was determined by PCR cycling and HRM analysis performed on the Rotor-Gene 6000™ (Corbett Research, Mortlake, Australia). The intercalating dye used was SYTO 9 (Invitrogen Life Technologies, Carlsbad, CA, USA). PCR assays were performed as previously described. All HRM reactions were run in triplicate [32].
Sequencing analysis was performed whenever an aberrant melting profile was obtained (ABI Prism 3130 sequencer). The sample’s DNA sequence was compared with reference sequences for detection of V600E Braf mutation.
Statistical analysis
Categorical variables were presented as counts and corresponding percentages, while continuous variables as means, standard deviations and respective ranges. Possible associations among categorical variables, either markers or clinico-pathological / treatment characteristics, were examined by the use of Fisher’s exact test. The Kruskal-Wallis test was used in order to examine for associations among categorical and continuous variables (i.e., age, time to bevacizumab discontinuation).
Overall survival (OS) was measured from the date of diagnosis until death from any cause or date of last contact. Respectively, overall survival-bevacizumab (OS-bevacizumab) was measured from the date of bevacizumab initiation, while time to bevacizumab discontinuation from the date of treatment initiation until discontinuation. Time-to-event distributions were estimated using Kaplan-Meier method, while the log-rank test was used to assess differences. In addition, univariate and multivariate Cox regression analysis with line of treatment as a time-dependent covariate was performed in order to adjust for the lead time bias caused by the fact that only those patients who survive long enough have the chance to receive treatment with bevacizumab as 2nd or 3rd line.
All univariate tests were two-sided and significance level was set at 5%. The statistical analysis was performed using the SAS software (SAS for Windows, version 9.3, SAS Institute Inc., Cary, NC).
In our analysis, we included 47 patients with primary brain tumors with median age 47.9 (from 21 to 69) years old. Original histologic diagnosis was glioblastoma multiforme in 35 cases while 12 patients had other glioma of low grade. 5 of the latter developed glioblastoma in subsequent biopsies. Radiotherapy was performed in every patient included. Maximal safe resection was achieved in 27 patients (57%), while partial resection or tumor biopsy was performed in 20 patients (43%). (Table 1)
Table 1. Patients Characteristics. 1: First Line Treatment, 2: Second Line Treatment, 3: Beyond Progression. A II: Astrocytoma Grade II, AA III: Anaplastic Astrocytoma Grade III, AOA III: Anaplastic Ologoastrocytoma Grade III, GBM: Glioblastoma, ODG II Oligodendroglioma Grade II, AO III: Anaplastic Oligodendroglioma Grade III.
All Patients |
N |
47 |
Age
|
Mean (SD) |
47.9 (12.2) |
Min-Max |
21-69 |
Original Histological Diagnosis |
A II |
2 (4.2%) |
AA III |
3 (6.4%) |
AOA III |
1 (2.2%) |
GBM |
35 (74.4%) |
ODG II |
3 (6.4%) |
ΑΟ ΙΙΙ |
3 (6.4%) |
Second Histological Diagnosis |
GBM |
5 (10.6%) |
AOA III |
2 (2.2%)5/ |
Lines of Treatment |
1 |
4 (8.6%) |
1, 2 |
9 (19.2%) |
1, 2, 3 |
2 (4.2%) |
2 |
24 (51%) |
2,3 |
2(4.2%) |
3 |
6 (12.8%) |
RT |
Yes |
47 (100%) |
Sex |
Female |
17 (36.2%) |
Male |
30 (63.8%) |
Type of Surgery |
Biopsy/Subtotal Resection |
20 (42.6%) |
Resection |
27 (57.4%) |
IDH1 was mutated in 13 cases (27.6%). IDH2 was not mutated in any case studied and in 3 patient samples the material was not enough for IDH mutation screening.
MGMT promoter was found to be methylated in 30 cases (63.8%), unmethylated in 13 cases (27.7%) and 4 samples were not analyzed.
EGFRvIII expression was seen in 11 patients’ tumors (23.4%). This variant was not present in 35 cases (74.5%) and 1 sample was not analyzed.
BRAF V600E mutation was present in 2 samples (4.3%) and the rest 45 samples (95.7%) were wild type. (Table 2)
Table 2. Biomarkers Frequency.
BIOMARKER |
Frequencies |
N |
% |
IDH1 |
31 |
66 |
normal |
mut |
13 |
27.7 |
not done |
3 |
6.4 |
IDH2 |
42 |
89.4 |
normal |
not done |
5 |
10.6 |
MGMT |
13 |
27.7 |
unmeth |
meth |
30 |
63.8 |
not done |
4 |
8.5 |
B-RAF |
45 |
95.7 |
normal |
mut |
2 |
4.3 |
EGFR VIII |
35 |
74.5 |
normal |
vIII variant |
11 |
23.4 |
not done |
1 |
2.1 |
Bevacizumab was prescribed as a monotherapy in 11 patients (23.4%) and in combination with other agents in 36 cases (76.6%). Bevacizumab was prescribed as a first line treatment with temozolomide after radiotherapy, in 15 patients (31%). Thirty-seven patients (78.7%) received bevacizumab as a second line treatment, in several combinations and as a third line treatment in 10 patients.
We also tried to investigate the statistical association of the biomarkers tested with the duration of therapy with bevacizumab. Patients received bevacizumab for 2 to 249 weeks. No statistically significant association between duration of therapy and biomarkers was found.
As expected, in our series overall survival was longer in patients with histology other than glioblastoma (median OS 221 vs. 127 weeks, log-rank p = 0.0043 and patients who had gross total resection as opposed to biopsy only or subtotal resection (median OS 154 vs. 93 weeks, p = 0.0381). IDH1 mutated patients and patients without EGFRvIII had longer overall survival (median OS 221 vs. 120 weeks, p = 0.0015 and median OS 151 vs. 120 weeks, p = 0.0372 respectively). When we studied only the cases with original histology of glioblastoma, there was no statistical association of overall survival and any biomarker analyzed. The same was true for overall survival from the date of bevacizumab initiation. (Table 3, 4 and 5)
Table 3. Overall Survival.
| |
Range |
|
95% CI |
|
Variables |
|
N |
Min |
Max |
Median |
LL |
UL |
P-value |
AAAll patients |
1 |
47 |
32 |
599 |
129 |
98 |
160 |
. |
Age cut off at 50% |
High |
23 |
40 |
483 |
98 |
62 |
129 |
0.004 |
| |
Low |
24 |
32 |
599 |
174 |
127 |
222 |
|
B-RAF |
Mut |
2 |
129 |
292 |
210 |
129 |
292 |
0.7231 |
| |
Normal |
45 |
32 |
599 |
127 |
98 |
160 |
|
EGFR VIII |
Normal |
35 |
52 |
599 |
151 |
114 |
186 |
0.0372 |
| |
vIII variant |
11 |
40 |
213 |
120 |
55 |
143 |
|
Hist. Diagnosis |
AA III-A II |
12 |
32 |
599 |
221 |
54 |
. |
0.0043 |
| |
GMB |
35 |
51 |
417 |
127 |
91 |
146 |
|
IDH1 |
Mut |
13 |
32 |
599 |
221 |
120 |
. |
0.0015 |
| |
Normal |
31 |
40 |
417 |
120 |
66 |
146 |
|
IDH2 |
Normal |
42 |
32 |
599 |
129 |
98 |
174 |
. |
MGMT |
Meth |
30 |
52 |
483 |
129 |
93 |
186 |
0.7004 |
| |
Unmeth |
13 |
40 |
599 |
154 |
55 |
179 |
|
Origi. Hist. Diagn |
A II |
2 |
133 |
599 |
. |
. |
. |
0.0004 |
| |
AA III |
3 |
40 |
120 |
54 |
40 |
120 |
|
| |
AOA III |
1 |
483 |
483 |
483 |
. |
. |
|
| |
GMB |
35 |
51 |
417 |
127 |
91 |
146 |
|
| |
ODG II |
3 |
221 |
415 |
. |
221 |
. |
|
| |
ΑΟ ΙΙΙ |
3 |
32 |
175 |
. |
151 |
. |
|
RT |
Yes |
47 |
32 |
599 |
129 |
98 |
160 |
. |
SEX |
FEMALE |
17 |
40 |
415 |
127 |
55 |
146 |
0.2575 |
| |
MALE |
30 |
32 |
599 |
151 |
98 |
186 |
|
type of surgery |
biopsy/subtotal resection |
20 |
32 |
292 |
93 |
66 |
151 |
0.0381 |
| |
Resection |
27 |
51 |
599 |
154 |
120 |
213 |
|
Table 4. Overall Survival in GBM Patients.
| |
Range |
|
95% CI |
|
Variables |
|
N |
Min |
Max |
Median |
LL |
UL |
P-value |
AAAll patients |
1 |
35 |
51 |
417 |
127 |
91 |
146 |
. |
Age cut off at 50% |
High |
19 |
51 |
186 |
98 |
62 |
127 |
0.0137 |
| |
Low |
16 |
52 |
417 |
154 |
91 |
213 |
|
B-RAF |
mut |
2 |
129 |
292 |
210 |
129 |
292 |
0.3037 |
| |
normal |
33 |
51 |
417 |
126 |
89 |
146 |
|
EGFR VIII |
normal |
25 |
52 |
417 |
127 |
66 |
160 |
0.3946 |
| |
vIII variant |
10 |
51 |
213 |
120 |
55 |
143 |
|
IDH1 |
mut |
4 |
114 |
213 |
170 |
114 |
213 |
0.4569 |
| |
normal |
28 |
52 |
417 |
120 |
89 |
146 |
|
IDH1 or IDH2 |
mut |
4 |
114 |
213 |
170 |
114 |
213 |
0.5528 |
| |
normal |
28 |
52 |
417 |
120 |
89 |
146 |
|
| |
not done |
3 |
51 |
100 |
. |
. |
. |
|
IDH2 |
normal |
30 |
52 |
417 |
126 |
89 |
154 |
. |
MGMT |
meth |
23 |
52 |
417 |
126 |
91 |
174 |
0.252 |
| |
unmeth |
9 |
51 |
179 |
124 |
55 |
160 |
|
RT |
yes |
35 |
51 |
417 |
127 |
91 |
146 |
. |
SEX |
FEMALE |
14 |
51 |
222 |
127 |
55 |
146 |
0.2532 |
| |
MALE |
21 |
58 |
417 |
126 |
91 |
174 |
|
type of surgery |
biopsy/subtotal resection |
14 |
55 |
292 |
93 |
66 |
160 |
0.7053 |
| |
resection |
21 |
51 |
417 |
127 |
64 |
174 |
|
Table 5. Overall Survival in Patients Since Bevacizumab Initiation.
| |
Range |
|
95% CI |
|
Variables |
|
N |
Min |
Max |
Median |
LL |
UL |
P-value |
AAAll patients |
1 |
47 |
0 |
296 |
54 |
39 |
76 |
. |
Age cut off at 50% |
High |
23 |
3 |
113 |
50 |
35 |
56 |
0.0566 |
| |
Low |
24 |
0 |
296 |
76 |
26 |
138 |
|
B-RAF |
mut |
2 |
56 |
76 |
66 |
56 |
76 |
0.918 |
| |
normal |
45 |
0 |
296 |
51 |
35 |
77 |
|
EGFR VIII |
normal |
35 |
0 |
296 |
51 |
35 |
93 |
0.3574 |
| |
vIII variant |
11 |
3 |
81 |
54 |
15 |
77 |
|
Hist. Diagnosis |
AA III-A II |
12 |
0 |
254 |
64 |
7 |
. |
0.2178 |
| |
GMB |
35 |
3 |
296 |
54 |
43 |
76 |
|
IDH1 |
mut |
13 |
0 |
254 |
47 |
20 |
. |
0.2467 |
| |
normal |
31 |
5 |
296 |
54 |
35 |
76 |
|
IDH2 |
normal |
42 |
0 |
296 |
55 |
39 |
76 |
. |
MGMT |
meth |
30 |
0 |
296 |
74 |
43 |
81 |
0.4789 |
| |
unmeth |
13 |
3 |
140 |
35 |
19 |
. |
|
Origi. Hist. Diagn |
A II |
2 |
0 |
116 |
. |
. |
. |
< 0.0001 |
| |
AA III |
3 |
7 |
25 |
15 |
7 |
25 |
|
| |
AOA III |
1 |
20 |
20 |
20 |
. |
. |
|
| |
GMB |
35 |
3 |
296 |
54 |
43 |
76 |
|
| |
ODG II |
3 |
93 |
254 |
. |
93 |
. |
|
| |
ΑΟ ΙΙΙ |
3 |
5 |
104 |
. |
35 |
. |
|
RT |
yes |
47 |
0 |
296 |
54 |
39 |
76 |
. |
SEX |
FEMALE |
17 |
0 |
159 |
48 |
20 |
107 |
0.7036 |
| |
MALE |
30 |
5 |
296 |
56 |
35 |
76 |
|
type of surgery |
biopsy/subtotal resection |
20 |
5 |
159 |
50 |
25 |
76 |
0.0883 |
| |
resection |
27 |
0 |
296 |
56 |
43 |
113 |
|
Associations among the biological markers studied did not reveal any statistically significant result.
Our analysis revealed a trend towards improved overall survival in glioblastoma patients with poor prognosis and the use of bevacizumab. Overall survival in original glioblastoma diagnosis patients whose tumors carried unfavorable prognostic factors, such as EGFRvIII and unmethylated MGMT promoter, was similar to the survival of those with favorable prognostic factors. Overall survival in EGFR vIII and wild type EGFR patients was 120 and 127 weeks respectively, while for unmethylated MGMT promoter and methylated MGMT promoter was 124 and 126 weeks respectively. This trend was not observed for IDH mutations. (Table 4)
The line of first bevacizumab treatment was also tested as predictor of survival and in initial analysis it was found that patients receiving bevacizumab in the 3rd line for the first time had favorable OS although not statistically significant longer (figure 1). In order to adjust for lead time bias, bevacizumab line of treatment was also used as a time-dependent factor in univariate and multivariate analysis. OS was measured from diagnosis date, and not from bevacizumab initiation date, since waiting time had to be taken into account. This time the 3rd line results were in contrast to the initial analysis. Patients who received bevacizumab as 2nd and/or 3rd line of treatment had a statistically significant increased risk of death compared to those who received it as a 1st line (2nd and 3rd vs. 1st, HR= 5.546 p < 0.001, 3rd vs. 1st HR = 4.549 p = 0.22, 2nd vs. 1st HR = 9.737 p = 0.0027, 3rd vs. 2nd HR = 0.92 p = 0.913). The findings regarding 3rd and/or 2nd line vs 1st line remained statistically significant even when bevacizumab treatment line was adjusted to patients’ molecular markers status and type of surgery (p = 0.0006) (Table 6).
Table 6. Analysis of maximum likelihood estimates After adjustment with patient profile, 2nd and 3rd line is still associated with a statistically significant increase in risk: HR=3.885, p=0.0006, along with variant vIII EGFR and normal IDH1.1st line vs 2nd and 3rd.
Parameter
|
Hazard |
Ratio |
Line Status |
2 & 3 vs 1 |
3.885 |
EGFR |
Variant vIII vs. normal |
2.32 |
IDH1 |
mut vs. normal |
0.333 |
MGMT |
meth vs. unmeth |
0.721 |
Type of surgery |
biopsy/subtotal resection vs. resection |
1.744 |
2021 Copyright OAT. All rights reserv
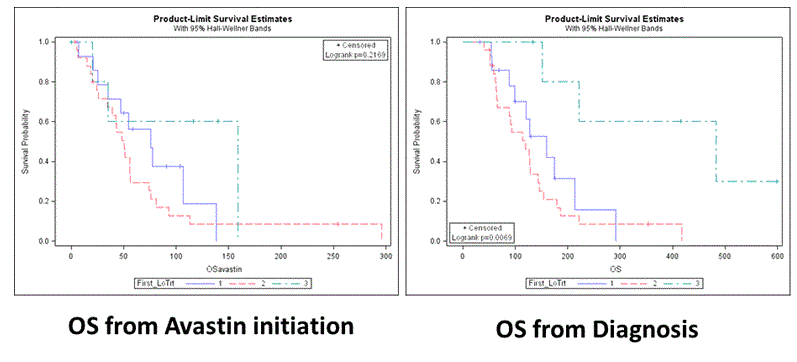
Figure 1. Kaplan Mayer OS.
Though the published prospective randomized studies with bevacizumab have been negative, all of us treating patients with high grade glioma have seen some patients derive clinical benefit. The need to identify the subpopulation that benefits is imperative, so that appropriately enriched studies can be designed.
As expected, our study showed statistically significant association of overall survival with glioblastoma histology. In the upcoming WHO classification, histopathology is combined with molecular and genomic classification of glioblastomas, in an attempt to better characterize their biological and clinical behavior [1,33-35]. Median overall survival of glioblastoma patients in our series was 2.5 years, quite a bit longer from the overall survival of one year reported in the literature. Radical surgical resection of patients’ tumors in conjunction with a very thorough and close monitoring and supportive care, are probably crucial determinants of this result. Reifenberger et al., investigated long-term survivors of glioblastoma with genome and transcriptome wide profiling, without finding any specific DNA copy number aberrations or expression signature. Besides the known molecular determinants like IDH mutations and MGMT methylation other factors, seem to be responsible for this long survival [35].
IDH1 mutated patients had longer survival (221 weeks) compared to normal IDH1 patients (120 weeks) (p value 0.0015). Favorable prognosis of IDH1 mutated patients is reported in several publications [13,14,35,36]. IDH1 mutation is not only a prognostic biomarker but also may be used as a target for immunotherapy [37].
Our study revealed a trend towards improved overall survival and the use of bevacizumab in glioblastoma patients with poor prognostic factors. For example, median overall survival since bevacizumab initiation in patients with EGFRvIII and unmethylated MGMT promoter was similar to those with favorable prognosis. This trend was not seen for IDH genes, probably because this mutation is crucial in gliomagenesis and its significance is thus unaffected.
Univariate analysis of the biomarkers tested as they relate to overall survival of patients treated with bevacizumab did not manage to reveal any statistically significant results therefore we did not proceed to multivariate analysis. Associations among the biological markers used in our study also did not yield any statistically significant result. Although the small number of patients in our study and its retrospective nature may be responsible for this result, other interpretations are also possible. One such factor may be intra-tumoral heterogeneity [38].
Bevacizumab treatment line was tested as predictor of survival and it was found that bevacizumab received in the 3rd line for the first time was associated with favorable OS although not with statistical significance. Taking into consideration that the time passed since bevacizumab initiation causes lead time bias, we further analyzed our data. Overall survival was measured from diagnosis date and not from bevacizumab initiation date, since waiting time had to be taken into account. Patients who received bevacizumab as a 2nd and/or 3rd line treatment demonstrated a considerably increased risk of earlier death, which indicates that higher line of treatment is associated with a shorter OS. This result remained statistically significant when adjusting for molecular parameters and surgery type.
Overall, we were unable to identify a marker that was associated with longer survival on bevacizumab. However, known poor prognosis parameters were not associated with worse outcome in our group. Despite the fact that our analysis has a small number of patients, and is thus more prone to random results, it is reasonable to assume that maybe this subgroup of glioblastoma patients that carry poor prognostic characteristics like IDH1 wild type, subtotal tumor resection and EGFRvIII expression may benefit from bevacizumab treatment.
The most likely explanation is that poor prognosis patients were more likely to get bevacizumab earlier, thus getting a comparative advantage, or a form of lead time advantage. This is shown by the fact that once line of therapy with bevacizumab is included in the assessment, the known poor prognosis parameters (such as EGFRvIII expression and wild type IDH) regain significance.
Whether the earlier initiation is in and of itself significant or it is the chance of getting bevacizumab in 2 or more lines, cannot be assessed since 1) most 11/15 patients that got bevacizumab in 1st line also got it in 2nd, 2) our numbers are too small and 3) there is no control group in the analysis, namely patients not treated with bevacizumab, therefore a causal relationship of this finding to bevacizumab per se cannot be proven.
Once again, we would like to reinforce that a plausible explanation for our findings may well be the retrospective nature of the study and small number of patients. An initial assumption that bevacizumab may benefit poor prognosis patients more, cannot be proven from this cohort. This is particularly the case since we have not included a non-bevacizumab treated cohort.
The assumption that earlier therapy with bevacizumab may in itself be more advantageous compared to therapy with subsequent lines is also not shown here due to the small numbers, and rather it was used to identify the lead time bias. Furthermore, this assumption is not supported by the published literature [6,7].
Both first and second line randomized control studies with bevacizumab in GBM have failed to show an advantage in the totality of the treated populations. However, all those involved in glioma patient care, have seen patients with impressive clinical responses. Therefore, the need to identify the subgroup that benefits is imperative, as regulatory constraints, are likely to deprive all patients from access to this agent.
The TCGA project subclassifies glioblastomas in 4 molecular types (classical, mesenchymal, neural and proneural) with specific genetic, epigenetic and transcriptional alterations [33]. Interestingly, Sandman et al., in their retrospective analysis of AVaglio raise the question whether molecular subtyping of glioblastoma tumors may reveal variants of this disease that may benefit with bevacizumab in first line [39]. The decision DX-GBM 9-gene assay separates patients with favorable outcome and proneural gene expression profile from those with poor prognosis expressing mesenchymal and angiogenesis genes, and was tested as a predictor for bevacizumab therapy in RTOG 0825 study [6,40]. This 9-Gene profile in combination with the existing clinical and molecular markers could be used to optimize therapeutic options for individual patients. Though, the application of the 9-Gene favorable predictive signature in patients with MGMT methylated glioblastomas treated with bevacizumab showed an adverse effect in survival, additional analysis of RTOG 0825 revealed a molecular signature of 43 genes and a 10-gene predictor of outcome for bevacizumab. The clinical application of this signature is under evaluation [41].
Our study implies that new treatment options in glioblastoma should be considered. There are a few hints that bevacizumab is a treatment that may work better in patients carrying poor prognostic factors. It is also obvious that the biomarkers tested here do not identify these glioblastoma patients adequately. New genetic analyses might reveal biological markers with direct relation to prognosis and treatment options.
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, et al. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97-109. [Crossref]
- Carlsson SK, Brothers SP, Wahlestedt C (2014) Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med 6: 1359-1370. [Crossref]
- Batchelor TT, Sorensen AG, Di Tomaso E, Zhang WT, Duda DG, et al. (2007) AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell 11: 83-95.
- Kreisl TN, Kim L, Moore K, Duic P, Royce C, et al. (2009) Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol 27: 740-745.
- Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, et al. (2009) Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol 27: 4733-40.
- Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, et al. (2014) A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 370: 699-708.
- Chinot OL, Wick W, Mason W, Henriksson R, Saran F, et al. (2014) Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 370: 709-722. [Crossref]
- Poulsen HS, Urup T, Michaelsen SR, Staberg M, Villingshoj M, et al. (2014) The impact of bevacizumab treatment on survival and quality of life in newly diagnosed glioblastoma patients. Cancer Manag Res 6: 373-387.
- Wick WAB, Gorlia T, Bendszus M, Sahm F, Taal W, et al. (2015) Phase III trial exploring the combination of bevacizumab and lomustine in patients with first recurrence of a glioblastoma: THE EORTC 26101 TRIAL. Neuro Oncol 17: (suppl 5).
- McNamara MG, Sahebjam S, Mason WP (2013) Emerging biomarkers in glioblastoma. Cancers (Basel) 5: 1103-1119. [Crossref]
- Michaelsen SR, Christensen IJ, Grunnet K, Stockhausen MT, Broholm H, et al. (2013) Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: an observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer 13: 402.
- Stupp R, Hegi ME, Mason WP, Van Den Bent MJ, Taphoorn MJ, et al. (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10: 459-466.
- Juratli TA, Kirsch M, Geiger K, Klink B, Leipnitz E, et al. (2012) The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. J Neurooncol 110: 325-333.
- Kalkan R, Atli EI, Ozdemir M, Ciftci E, Aydin HE, et al. (2015) IDH1 mutations is prognostic marker for primary glioblastoma multiforme but MGMT hypermethylation is not prognostic for primary glioblastoma multiforme. Gene 554: 81-86.
- Cominelli M, Grisanti S, Mazzoleni S, Branca C, Buttolo L, et al. (2015) EGFR amplified and overexpressing glioblastomas and association with better response to adjuvant metronomic temozolomide. J Natl Cancer Inst p. 107.
- Weller M, Kaulich K, Hentschel B, Felsberg J, Gramatzki D, et al. (2014) Assessment and prognostic significance of the epidermal growth factor receptor vIII mutation in glioblastoma patients treated with concurrent and adjuvant temozolomide radiochemotherapy. Int J Cancer 134: 2437-4247.
- Neagu MR, Reardon DA (2015a) Rindopepimut vaccine and bevacizumab combination therapy: improving survival rates in relapsed glioblastoma patients. Immunotherapy 7: 603-606.
- Neagu MR, Reardon DA (2015) An Update on the Role of Immunotherapy and Vaccine Strategies for Primary Brain Tumors. Curr Treat Options Oncol 16: 54. [Crossref]
- Schuster J, Lai RK, Recht LD, Reardon DA, Paleologos NA, et al. (2015) A phase II, multicenter trial of rindopepimut (CDX-110) in newly diagnosed glioblastoma: the ACT III study. Neuro Oncol 17: 854-861.
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M (2005) Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res 11: 1119-1128.
- Kannan K, Inagaki A, Silber J, Gorovets D, Zhang J, et al. (2012) Whole-Exome Sequencing Identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget 3: 1194-203.
- Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, Aldape KD, Yung WK, et al. (2015) Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372: 2481-2498.
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, et al. (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110: 6021-6026.
- Mosrati MA, Malmstrom A, Lysiak M, Krysztofiak A, Hallbeck M, et al. (2015) TERT promoter mutations and polymorphisms as prognostic factors in primary glioblastoma. Oncotarget 6: 16663-16673.
- Takahashi Y, Akahane T, Sawada T (2015) Adult classical glioblastoma with a BRAF V600E mutation. World J Surg Oncol 13: 100. [Crossref]
- Karsy M, Neil JA, Guan J, Mahan MA, Colman H, et al. (2015) A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus 38: E4.
- Jubb AM, Harris AL (2010) Biomarkers to predict the clinical efficacy of bevacizumab in cancer. Lancet Oncol 11: 1172-1183. [Crossref]
- Chen C, Huang R, Maclean A, Muzikansky A, Mukundan S, et al. (2013) Recurrent high-grade glioma treated with bevacizumab: prognostic value of MGMT methylation, EGFR status and pretreatment MRI in determining response and survival. J Neurooncol 115: 267-276.
- Van Den Bent MJ, Dubbink HJ, Marie Y, Brandes AA, Taphoorn MJ, et al. (2010) IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res 16: 1597-1604.
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, et al. (2005) Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012-2024.
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, et al. (2000) Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343: 1350-1354.
- Negru S, Papadopoulou E, Apessos A, Stanculeanu DL, Ciuleanu E, et al. (2014) KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study. BMJ Open 4: e004652.
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98-110.
- Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, et al. (2015) Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol 129: 679-693.
- Reifenberger G, Weber RG, Riehmer V, Kaulich K, Willscher E, et al. (2014) Molecular characterization of long-term survivors of glioblastoma using genome- and transcriptome-wide profiling. Int J Cancer 135: 1822-1831.
- Molenaar RJ, Verbaan D, Lamba S, Zanon C, Jeuken JW, et al. (2014) The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 16: 1263-1273. [Crossref]
- Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, et al. (2014) A vaccine targeting mutant IDH1 induces antitumour immunity. Nature 512: 324-327.
- Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR (2015) Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol 5: 55.
- Sandmann T, Bourgon R, Garcia J, Li C, Cloughesy T, et al. (2015) Patients with Proneural Glioblastoma May Derive Overall Survival Benefit from the Addition of Bevacizumab to First-Line Radiotherapy and Temozolomide: Retrospective Analysis of the AVAglio Trial. J Clin Oncol 33: 2735-2744.
- Colman H, Zhang L, Sulman EP, Mcdonald JM, Shooshtari NL, et al. (2010) A multigene predictor of outcome in glioblastoma. Neuro Oncol 12: 49-57.
- Sulman EWM, Blumenthal D (2013) Molecular predictors of outcome and response to Bevacizumab (BEV) based on analysis of RTOG 0825, a phase III trial comparing chemoradiation (CRT) with and without BEV in patients withh newly diagnosed glioblastoma (GBM). J Clin Oncol 31:18.
