Objetives: To describe and compare the protein profiles of tissue, serum and saliva from a patient who presented a typical case of oral lichen planus and from healthy controls.
Methods: We first analyzed the histological characteristics of a patient’s tissue sample with a typical case of oral lichen planus. Protein profiles obtained from oral mucosa tissue, serum and saliva were analyzed using two-dimensional (2-D) electrophoresis and further compared with similar samples obtained from healthy controls.
Results: (2-D) Protein profiles consisted of a total of 228 spots in the pathological tissue, 206 spots in the pathological saliva and 216 spots in pathological serum. We observed several differentially expressed proteins in the sample of the oral lichen planus in comparison to normal samples. Among these proteins, the precursor of fibrinogen and the salivary amylase were identified by mass spectrometry.
Conclusions: We detected several differential expressed proteins in oral lichen planus in comparison to healthy samples. Although this work shows experiments comparin a single patient versus samples of saliva and serum from their healthy controls, it could represent the future to find biomarkers in this setting and possibly any diseases. Finally, in the future will be necessary to make more experiments and identify other differentially expressed proteins in order to detect biomarkers of this disease.
Lichen planus, saliva, serum, mass spectrometry assay, proteomics
Oral lichen planus (OLP) is a chronic inflammatory disorder, relatively common of oral mucosa, which represents an autoimmune disease [1-2]. It could be presented in an isolated form or concomitantly with a cutaneous lichen planus [3]. Genital mucosal surfaces, scalp, and nails can be also affected [4]. OLP tends to be chronic, relapsing, and difficult to treat.
The presentation of the disease often occurs in the fourth decade of life and most often affects the female gender within a 4:1 rate. It could be present in about 0.5–2% of the world population and persisting up to more than 20 years without spontaneous remission [5]. Clinical variants include: reticular, papular, plaque, erosive, atrophic and blistering subtypes [6]. Severe morbidity is related to the erosive form [3].
Although, the detailed etiology and physiopathology is still unclear, it has been suggested that the inflammatory response in OLP is characterized and mediated by the accumulation and expansion of T-helper 1 (Th1) lymphocytes [1]. Several lines of evidence have been suggesting that a complex cytokine network plays an important role in the exacerbation and perpetuation of OLP [1,7].
The initial event is the antigen-specific presentation by basal keratinocytes to CD4+ lymphocytes, and the consequently attraction of lymphocytes to the basement membrane occurs due to cytokine-mediated regulation of adhesion molecules in endothelial cells. Macrophages, Langerhans cells, lymphocytes and surface keratinocytes are responsible for the regulation of TNF-α, IFN-γ and IL-1. In addition, mast cell degranulation releases pro-inflammatory mediators, such as TNF-α, chymase and tryptase. Chymase is a mast cell protease that activates MMP-9 and further enables the disruption of the basement membrane. These mechanisms may be combined to cause a T-cell accumulation in the superficial layer of the lamina propria, basement membrane disruption, intra-epithelial T-cell migration and keratinocyte apoptosis in OLP. Thus, a vicious cycle related to the chronic-inflammatory nature of the disease is established [8].
On the other hand, proteomics, proteomics, defined in its most broad terms, is the study of the proteome: the collection of all proteins expressed from the genome, in all isoforms, polymorphisms, and post-translational modifications under a specific condition [9]. Two-dimensional electrophoresis (2DE), combined with mass spectrometry (MS) have been widely used to analyze protein profiles, identify proteins with differential expression, and discover cancer biomarkers for the early diagnosis, prognosis, and prediction of the outcome of therapies [10]. However, despite the multiple applications of proteomics in cancer research, there are not similar reports on oral lichen planus research.
Even now, the scientific knowledge related with OLP has been focused in the pathological descriptions and immunological response. However, there is few information respect to the protein profile of OLP, including other samples as serum, and whole unstimulated saliva protein profiles from OLP patients.
The aim of this study was to describe a typical clinical case of OLP, to analyze the proteomic expression profile of the pathological tissue, as well as the whole unstimulated saliva and serum from the OLP patient and finally, to compare these protein profiles with those obtained from control samples in order to suggest possible biomarkers of OLP.
A 69-year-old female patient presented multiple intraoral lesions located bilaterally in tongue, buccal mucosa, floor of the mouth, and gums with a clinical evolution of three months (Figure 1). According to clinical classification [5], and the definition of OLP by the World Health Organization, reticular and plaque subtypes alternating with erythematous/ ulcerative subtype were observed (Figure 1). Low pain symptom was related. The personal pathological history revealed: hepatitis type B suffered at 6-years-old. However, Hepatitis B serologic test was performed at 51-years-old with a negative result. Since 1997, OLP was diagnosed with three to four periods of remission and exacerbation. The patient underwent myocardial infarction at 62-years-old, treated with a coronary stent implantation and acetylsalicylic acid, which was suspended one week before the biopsies were taken. Hypercholesterolemia was treated with atorvastatin calcium. Four months before symptoms of GERD (gastroesophageal reflux disease) were presented, insomnia caused by emotional problems was treated with clonazepam and alprazolam.
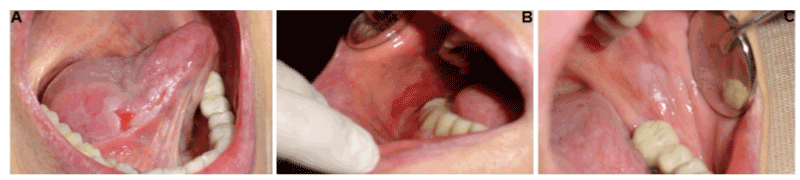
Figure 1. Oral manifestations in a patient with oral lichen planus. A) Right lateral border of the tongue. Plaque subtype alternating with ulcerative subtype was diagnosed. B) On right buccal mucosa, erythematous/ ulcerative subtype was clinically detected. C) On left buccal mucosa, plaque subtype was observed.
The patient presented periods of exacerbation and remission of OLP lesions (Figure 2). Clinical monitoring for three years has been given to the patient, after the therapeutic prescription of fluocinonide, triamcinolone, prednisolone and tacrolimus.
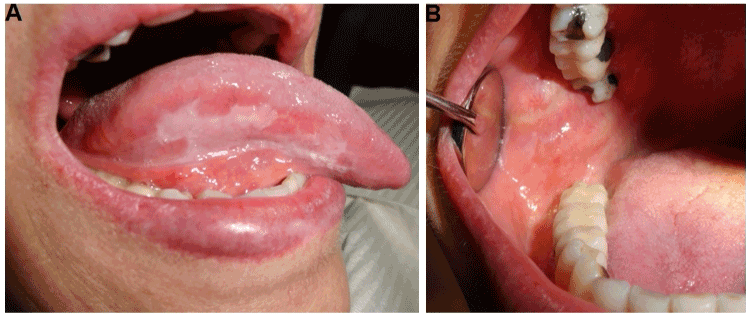
Figure 2. Clinical manifestations of OLP patient after 3 years follow-up. A) Right lateral border of the tongue. B) Right buccal mucosa showed a remission and an asymptomatic period.
Samples
The patient lesions with a presumptive OLP diagnostic were observed in a private dental clinic. Then, patient was referred to the Department of Maxillofacial Surgery of the Hospital Juárez de México. Two incisional biopsies were carried out, one from lateral right tongue with reticular pattern and other from right buccal mucosa showing erosive pattern.
For proteomics, this patient with OLP and four control cases were used. At the same session, the samples of saliva and serum were set on sample buffer before the biopsies were taken.
The present study followed the Declaration of Helsinki for the medical protocol and ethics, and the institutional committee of research and ethics approved the study (registration no. HJM 1996/11.03.08).
Samples preparation
Tissue samples (both pathological and control tissues) were extracted from the patient during biopsy procedures. Then, they were cleaned using physiological solution, frozen in liquid nitrogen, and stored at -70oC until use. Whole unstimulated saliva and blood samples were collected from the patient before surgery and from healthy controls. Saliva samples were filtered using 0.22µm disc nylon syringe and complete protease inhibitor cocktail ™ (Roche, Germany) was added, frozen, and stored at -20°C until use.
Histopathology
To analyze the morphological aspects of the samples, these were fixed in 10% buffered formaldehyde, sectioned, mounted on microscope slides, stained with hematoxylin and eosin [11] and examined by optical microscopy (Nikon H550 L, Yokohama, Japan). Later, samples were reviewed in at least 5 high-powered fields by two qualified oral and maxillofacial pathologists (Universidad Tecnológica de México, Mexico). Finally, the slides were selected according to the histopathological classification of World Health Organization [12].
Protein extraction
Protein extraction from tissue, samples were rinsed in physiological solution, frozen in liquid nitrogen, mechanically pulverized and suspended [200 mg tissue/mL] in sample buffer (7 M urea, 2 M thiourea, 4% CHAPS, 2% IPG buffer, 40 mM DTT) containing complete protease inhibitor cocktail™ (Roche, Germany). Then, samples were disrupted by sonication. Insoluble material was removed by centrifugation (20,000 xg for 5 min. at 4 ºC), and the supernatant was preserved. Additionally, proteins were precipitated with acetone-TCA and the 2D Clean-Up Kit (Amersham Biosciences, USA). The precipitates were dissolved in rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 0.5%, IPG buffer and 0.1% bromophenol blue) supplemented with 2 mM DTT. Protein concentration was measured using 2D Quant Kit (Amersham Biosciences, USA) according to the manufacturer’s recommendations.
Serum samples
The sera were obtained by centrifugation of the blood (2500 xg for 5 min at 4 °C). Then, 100 mg serum/mL was added in sample buffer containing the complete protease inhibitor cocktail. Insoluble material was removed by centrifugation (20,000 xg for 5 min at 4°C), and the supernatant was preserved. Additionally, proteins were precipitated twice with acetone-TCA. Then, serum samples [100 mg/mL] and 600 µL of methanol were added and mixed, later 150 ml of chloroform was added and mixed, finally 450 µL of Milli-Q water was added and mixed. Insoluble material was removed by centrifugation (20,000 xg for 5 min. at 4ºC), and the white disk between phases was preserved. Afterwards, 450 µL of methanol was added and mixed, centrifuged at 20,000 xg for 5 min at 4°C, mixed with Vortex and centrifuged (20,000 xg for 5 min. at 4ºC). Finally, each precipitate was diluted in rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG buffer and 0.1% bromophenol blue) supplemented with 2 mM DTT. Protein concentrations were measured using 2D Quant Kit (Amersham Biosciences, USA) according to the manufacturer’s recommendations.
Saliva samples
Each saliva sample [100 mg saliva/mL] and 600 µL of methanol were added and mixed, later 150 µL of chloroform was added and mixed, and finally 450 ml of Milli-Q water was added and mixed. Insoluble material was removed by centrifugation (20,000 xg for 5 min. at 4ºC), and the white disk between phase was preserved. Later, 450 µL of methanol was added and mixed, then centrifuged at 20,000 xg for 5 min at 4 °C, mixed and centrifuged (20,000 xg for 5 min at 4 ºC). Finally, each precipitate was diluted in rehydration solution (7 M urea, 2 M thiourea, 2% CHAPS, 0.5% IPG buffer and 0.1% bromophenol blue) supplemented with 2 mM DTT. Protein concentrations were measured using 2D Quant Kit (Amersham Biosciences, USA) according to the manufacturer’s recommendations.
One-dimensional electrophoresis (1-DE)
To demonstrate the quality of the proteins obtained, 35 μg of proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE 10%) stained with Coomassie Blue [15].
Two-dimensional electrophoresis (2-DE)
To obtain the protein profile of the pathological samples, but also to analyze its differential expression with respect to controls, we performed 2-DE of wide range ampholyte pH 3–10. The profiles were further visualized by colloidal Coomassie Blue G-250 staining. To minimize analytical (gel-gel) variation, two-dimensional gels for each sample were made.
This protocol was applied to tissue, saliva, and serum samples. Protein extracts (160 µg) suspended in rehydration solution (125 μL) were used to rehydrate Immobiline Drystrip Gels, pH 3-10 of 7 cm (GE Healthcare Life Science) for 18 h at room temperature. Isoelectric focusing was performed in an Ettan IPGphor 3 Isoelectric Focusing System (GE Healthcare, USA) at 16-20 kVh for 5 h. Then, the immobilized pH gradient (IPG) strips were incubated for 10 min in reducing and alkylating 2-DE equilibration buffer (6 M urea, 75 mM Tris-HCl, pH 8.8, 29.3% glycerol, 2% sodium dodecyl sulfate and 0.1% bromophenol blue) plus 65 mM DTT and 135 mM iodoacetamide, successively. For SDS polyacrylamide gel electrophoresis (SDS-PAGE), a standard vertical electrophoresis system was used with 10% polyacrylamide gels (8 cm × 8cm) in a Gibco BRL V16 gel system. Gels were stained with Colloidal Coomassie Blue G-250 (Bio-Safe Coomassie Stain, Bio-Rad Laboratories, USA). A digital image of the gels was obtained using scanning densitometry (Image Scanner, Amersham Biosciences, USA) and analyzed with Image Master 2D Platinum software, version 7.0 (GE Healthcare Life Sciences, Switzerland).
Mass spectrometry and identity analysis
Two spots present only in the pathological samples were excised, then subjected to in-gel trypsin duplicate digestion and analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Peptide mass fingerprinting and MS/MS data were searched against the human genome database using the MASCOT, program version 2.1 (available at: http://www.matrixscience.com) allowing a monoisotopic mass tolerance of 1 Da. Methionine oxidation and 1 missed trypsin cleavage were used during the database search.
Pathological features of OLP
In the tissue obtained from the tongue with acanthotic epithelium we observed thickened rete peg with saw tooth appearance and atrophic epithelium areas. In the underlying connective tissue, a band of lymphocytic inflammatory infiltrate with sub-basal and parabasal location was observed (Figure 3A). Liquefaction of the basement membrane, vacuolization and destruction of epithelial basal cells, and basal apoptotic cells with sub-epithelial separation were recognized (Figure 3B).
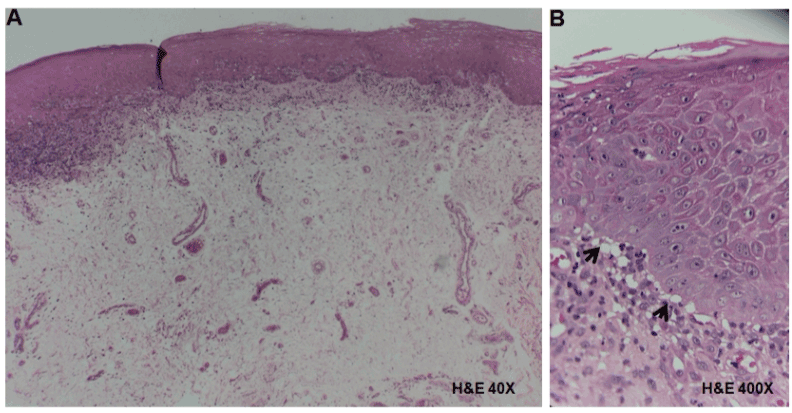
Figure 3. Histopathological analysis of tissue obtained from the tongue in a patient with OLP. A) Hyperplastic epithelium showing acanthosis, and in the underlying connective tissue a band of lymphocytic inflammatory infiltrate in a sub-basal localization (H&E, 40X). B) Liquefaction of the basement membrane, vacuolization and destruction of basal cells (arrows) with sub-epithelial separation (H&E, 400X).
In the tissue obtained from the buccal mucosa, an ulcerative area was observed (Figure 4A). In addition, this lesion showed atrophic epithelium, dense lymphocytic inflammatory infiltrate in the lamina propria and also vascular tissue hyperemia (Figure 4B). Thus, the microscopic findings confirmed the OLP diagnosis.
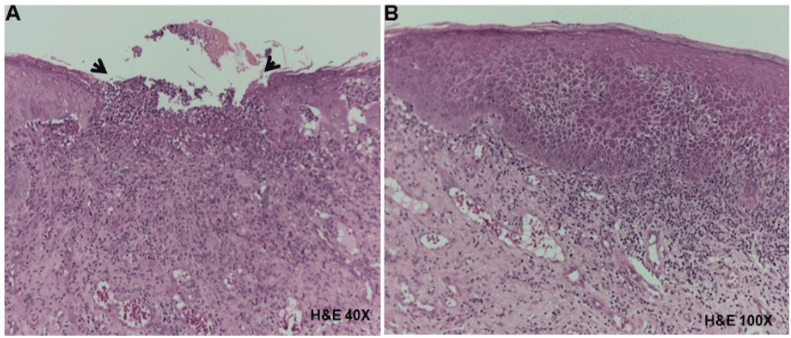
Figure 4. Histopathological analysis of tissue obtained from the right buccal mucosa in a patient with OLP. A) Ulcerative area (arrows) showing connective tissue exposition (H&E, 40X). B) Hyperplasic and atrophic epithelium, dense lymphocytic inflammatory infiltrate in the lamina propria and vascular hyperemia (H&E, 400X).
Proteomic profiles of OLP
Two-dimensional electrophoresis gels revealed the protein profiles of salivary, serum and pathological tissue. Tissue and serum from pathological and control samples displayed similar protein profiles for each tissue, with more than 100 protein spots (Table 1) showing molecular masses ranging from >250 to 5 kDa and pI values between 3 and 10. However, several protein spots consistently displayed significant differences in expression between pathological and healthy samples. Representative 2 DE of pathological and control sample are shown (Figure 5, 6). Interestingly, the protein profiles of saliva from patient and controls were quite different (Figure 7).
Table 1. Identification of overexpressed proteins from control and oral lichen planus samples
| |
|
|
Sample |
TS |
AS |
Control tissue |
169 |
|
Pathological tissue |
228 |
|
Control saliva |
81 |
|
Pathological saliva |
206 |
1 |
Control serum |
104 |
|
Pathological serum |
216 |
1 |
TS: Total spots obtained in 2-DE gels. AS: Analyzed spots using LC-MS/MS. |
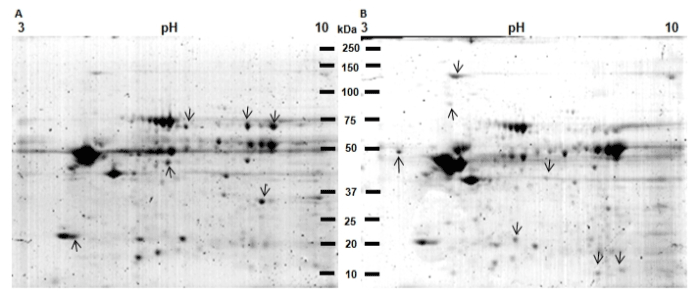
Figure 5. 2DE protein profiles of control tissue (CT) and pathological tissue (PT) of the patient with OLP. A) Protein profile of CT. B) Protein profile of PT. Proteins from TC and PT were extracted and separated in 2DE. Gels were stained with Colloidal Coomassie Blue G-250. Proteins differentially expressed between both samples are labeled (arrows). The CT showed 169 spots (A) and the pathological tissue from the OLP patient 228 spots (B).
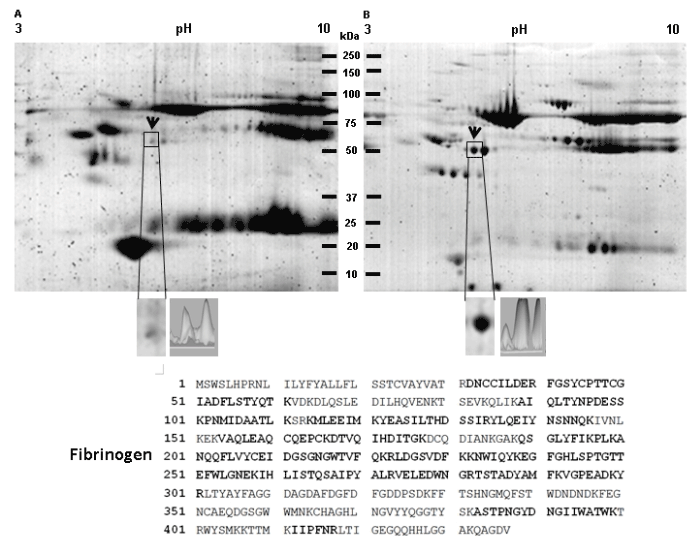
Figure 6. Identification of fibrinogen overexpression in a 2D protein profiling of serum from a patient with oral lichen planus. A) Protein profile of serum from healthy control patient. B) Protein profile of serum from a patient with oral lichen planus. Proteins from both samples were extracted and separated in 2DE. Gels were stained with Colloidal Coomassie Blue G-250. Proteins differentially expressed between both samples are labeled (arrows). The spot that consistently showed significant up regulation in serum from a patient with oral lichen planus is indicated by a frame. C) Magnification of the spot surrounded by a square and a differential intensity analysis is shown. The spot surrounded by a square was excised from gels and subsequently analyzed by LC-MS/MS. MS/MS analysis identified the differential spot as a fibrinogen precursor, which amino acid sequence is shown. The peptides identified by LC-MS/MS are enlarged and bold. Finally, the control serum showed 104spots (A) and the serum from patient showed 216 spots (B).
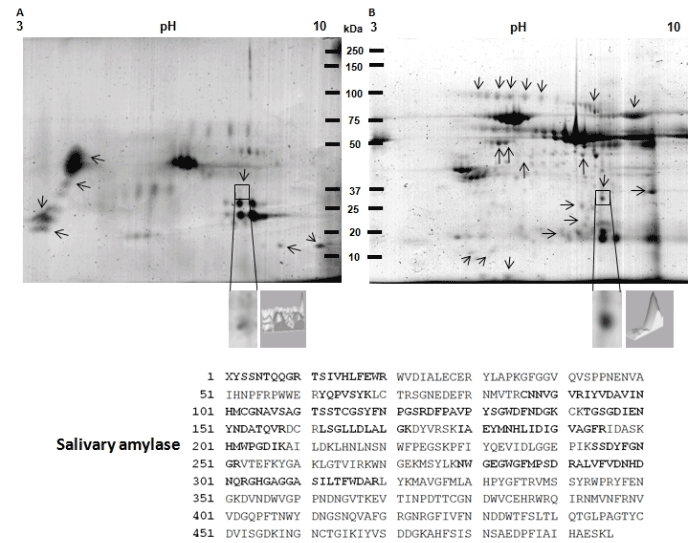
Figure 7. Identification of salivary amylase overexpression in a 2D protein profiling of saliva from patient with oral lichen planus. A) Protein profile of saliva from healthy control patient. B) Protein profile of saliva from patient with oral lichen planus. Proteins from both samples were extracted and separated in 2DE. Then, gels were stained with Colloidal Coomassie Blue G-250. Proteins differentially expressed between both samples are labeled (arrows) and one of the spots that consistently showed significant up-regulation in saliva from patient with oral lichen planus is indicated by a frame. The spot that consistently showed significant up regulation in serum from a patient with oral lichen planus is indicated by a frame. C) Magnification of the spot surrounded by a square and a differential intensity analysis is shown. The spot surrounded by a square was excised from gels and subsequently analyzed by LC-MS/MS. MS/MS analysis identified the differential spot as the salivary amylase, which amino acids sequence is shown. The peptides identified by LC-MS/MS are enlarged and bold. Finally, the control saliva showed 81 spots (A) and the saliva from patient showed 206 spots (B).
Fibrinogen precursor and salivary amylase are overexpressed in OLP patient samples
From gels of saliva and serum, two spots differentially expressed in the OLP samples were chosen for the mass spectrometry analysis (Figure 6-7). These spots were selected among the spots with greater overexpression in the pathological samples compared with their control.
The spot selected in serum sample (Figure 6) showed a moyilecular weight of 51 kDa and a pI of 5.3 (Figure 6, lined squared spot). The spectrometry mass analysis indicated that this protein spot corresponds to the precursor of “fibrinogen” (Table 2). The spot obtained from whole unstimulated saliva corresponds to the salivary amylase (chain A) (Table 2). This protein spot showed a molecular weight of 35 kDa and pI 8.0 (Figure 7, lined squared spot).
Table 2. Identification of proteins with differential expression in oral lichen planus samples
Sample |
NCBI No. |
Name |
Theoretical Protein
ID Mass
KDa/pI |
Score |
Sequence
Coverage |
Major functions |
Serum |
gi 6650830 |
Fibrinogen gamma chain isoform gamma-A precursor |
50103/5.7 |
1693 |
56% |
Fibrinogen is involved in blood clotting, being activated by thrombin to assemble into fibrin clots. |
Saliva |
gi 27065641 |
Chain A, human salivary amylase |
56495/6.21 |
3273 |
37% |
It has the role of a mobile loop in substrate binding and enzyme activity of human salivary amylase. |
Accession numbers are from the MASCOT database.
Oral lichen planus is an autoimmune disease mediated by chronic inflammation, commonly expressed in the oral mucosa [3]. The detailed inflammatory mechanism in OLP and the T lymphocyte-mediated immune response remain unclear [4,5].
In the present study, we analyzed the profile of proteins expressed from the pathological tissue in a patient with OLP and also the profiles of proteins detected in serum and saliva from the same patient. Moreover, we compared protein profiles of pathological tissue with healthy tissue from the same patient. The saliva and serum proteins profiles were compared using samples from healthy persons.
Once obtained the protein profile analysis we detected similar patterns in the pathological tissue, serum and saliva of the patient with OLP compared with the control samples.
However, we recognized different spots differentially expressed (Figure 5-7) among samples. As in many other conditions, e.g. cancer, gene and protein expression are altered, modifying cellular and molecular processes leading to disease progression or to chronic condition, among others. In our understanding, this is the first report examining the proteome expression profiles considering the analysis of tissue, serum, and saliva of a patient with OLP and healthy samples.
The obtained results in this report might be useful for future studies, as the complete protein profiles have not been shown in previous published scientific reports. However, we consider that our results are not conclusive due to the fact that the patient case reported in this study had a large clinical and pharmacological history. In addition, a higher number of patients and samples replications may contribute to the better understanding of the proteomic profile in OLP.
The first selected spot identified as a precursor of fibrinogen belongs to OLP patient’s serum. In addition, such spot was found in the visual analysis also in the pathological tissue and saliva of the OLP patient. The second spot detected in saliva, identified as salivary amylase (chain A) was chosen due to apparent exclusive expression in this fluid. The implications of identifying new potential non-invasive or minimal-invasive biomarkers are highly important. Saliva and even serum available biomarkers might be a potentially helpful in early diagnosis, monitoring disease progression, or treatment responses for minimally trained personnel [16]. These biological fluids are easily obtainable and can give useful information both in systemic and oral diseases [17,18].
Unstimulated saliva is believed to represent an equilibrated condition, having less influence from salivary glands [19].
Although there are previous analyses reporting until 2290 proteins founded in whole saliva and approximately 27% of the whole-saliva proteins are found in plasma [18]. In our experiment we found 206 spots in the saliva and 216 in the serum of the patient with OLP. This project was performed in order to identify at least one biomarker in saliva and/or serum. The obtaining of a precursor of fibrinogen as a biomarker candidate may have important implications, as it was found in all pathological samples probed here. This potentially expressed biomarker remains to be validated in more patients and in large scale.
There is only one scientific report which shows a total of 31 protein spots representing 14 proteins with at least two-fold difference in abundance between OLP and control samples. They suggested new biomarkers which may play a role in inflammation and immune response of OLP [20].
The precise role of gamma chain isoform gamma-A precursor of fibrinogen in OLP pathogenesis may be related with the chronic inflammatory process and the repair response of the oral mucosa. Fibrinogen is involved in blood clotting, being activated by thrombin to assemble into fibrin clots. It remains to be investigated the precise implication of this protein in the physiopathology of the disease.
We observed many differentially expressed proteins and we believe that some of these could eventually be used as biomarkers of OLP in serum or saliva. However, it is not known whether these proteins are generated as a result of OLP disease or as a result of the changes caused by the drugs that the patient is consuming. Among spots identified to express differentially, the fibrinogen was considerably important, not only for its function but also for its expression in all pathological samples analyzed. Fibrinogen overexpression in pathological tissue and serum of OLP patients could be considered normal because the oral mucosa is constantly under immunological and inflammatory challenges, which involve this molecule. However, fibrinogen overexpression in the saliva of the OLP patient was an unexpected result. The salivary amylase protein is considered to have a normal role in the oral cavity. The overexpression of amylase protein in OLP patient needs to be further validated. Finally, we consider necessary to further analyze more samples and evaluate the profile of differentially expressed proteins in OLP patients. It remains to be investigated the implications of differentially expressed proteins in the biological process of OLP disease and the use of such proteins as new specific biomarkers.
The authors declare no conflict of interest.
We appreciate the technical assistance provided by Leticia Cortés-Martínez and the support provided by the programme: ''PAPCA: FESI-DIP-PAPCA-2016-8.
- Pekiner FN, Demirel GY, Borahan MO, Ozbayrak S (2012) Cytokine profiles in serum of patients with oral lichen planus. Cytokine 60: 701-706.
- Buajeeb W, Okuma N, Thanakun S, Laothumthut T (2015) Direct Immunofluorescence inOral Lichen Planus. J Clin Diag Res 9: ZC34-347.
- Nico MM, Fernandes JD, Lourenço SV (2011) Oral lichen planus. Anais Brasileiros de Dermatologia 86(4): 633-641. [Crossref]
- Crincoli V, Di Bisceglie MB, Scivetti M, Lucchese A, Tecco S, et al. (2011) Oral lichen planus: update on etiopathogenesis, diagnosis and treatment. Immunopharmacol Immunotoxicol 33: 11-20.
- Al-Hashimi I, Schifter M, Lockhart PB, Wray D, Brennan M, et al. (2007) Oral lichenplanus and oral lichenoidlesions: diagnostic and therapeuticconsiderations. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology 103 Suppl: S25.e1-12. 29.
- Schlosser BJ (2010) Lichen planus and lichenoid reactions of the oral mucosa. Dermatology and Therapy 23: 251-267. [Crossref]
- Tao XA, Li CY, Rhodus NL, Xia J, Yang XP, Cheng B (2008) Simultaneous detection of IFN-gamma and IL-4 in lesional tissues and whole unstimulated saliva from patients with oral lichen planus. J Oral Pathol Med 7: 83-87.
- Roopashree MR, Gondhalekar RV, Shashikanth MC, George J, Thippeswamy SH, et al. (2010) Pathogenesis of oral lichen planus--a review. J Oral Pathol Med 39: 729-734.
- Graham DR, Elliott ST, VanEyk JE (2005) Broad-based proteomic strategies: A practical guide to proteomics and functional screening. The J Physiol 563:1.
- Guo S, Zou J, Wang G (2013) Advances in the proteomic discovery of novel therapeutic targets in cancer. J Drug Desi Develop Ther 7:1259.
- Kamath KP, Vidya M, Shetty N (2010) Nucleolar organizing regions and alpha-smooth muscle actin expression in a case of ameloblastic carcinoma. HeadNeck Pathol 4:157.
- Van der Waal I (2009) Oral lichen planus and oral lichenoid lesions; a critical appraisal with emphasis on the diagnostic aspects. Oral Surg Oral Med Oral Pathol Oral Radiol 14: 310-314.
- Gorg A, Obermaier C, Boguth G, Harder A, Scheibe B, et al. (2000) The currunt state of two-dimensional electrophoresis with immnobilized pH gradients. Electrophoresis 21: 1037-1053.
- Pérez E, Gallegos JL, Cortés L, Calderón KG, Luna JC, et al. (2010) Identification of latexin by a proteomic analysisin rat normal articular cartilage. Proteome Sci 8: 27.
- García-Muñoz A, Rodríguez MA, Bologna-Molina R, Cázares-Raga FE, Hernández- Hernández FC, et al. (2012) The orosomucoid 1 protein (α1 acid glycoprotein) is overexpressed in odontogenic myxoma. Proteome Sci 10: 49.
- Al-Tarawneh SK, Border MB, Dibble CF, Bencharit S (2011) Defining salivary biomarkers using mass spectrometry-based proteomics: a systematic review. OMICS 15: 353-361.
- Küçükkolbaşi H, Küçükkolbaşi S, Dursun R, Ayyildiz F, Kara H (2011) Determination of defensin HNP-1 in human saliva of patients with oral mucosal diseases. J Immunoassay Immunochem 32: 284-295.
- Loo JA, Yan W, Ramachandran P, Wong DT (2010) Comparative human salivary and plasma proteomes. J Dent Res 89: 1016–1023. [Crossref]
- Ohshiro K, Rosenthal DI, Koomen JM, Streckfus CF, Chambers M, et al. (2007) Pre-analytic saliva processing affects proteomic results. And biomarker screening of head and neck squamous carcinoma. Int J Oncol 30: 743-749.
- Yang LL, Liu XQ, Liu W, Cheg B, Li MT (2006) Comparative analysis of whole saliva proteomes for the screening of biomarkers for oral lichen planus. Inflammat Res 55: 405-407.







