Acetylation is one of protein modifications and important for gene expression by modulating activity or conformation of transcription factors and histones. Although Bromodomain-containing proteins which bind to acetylated histones have critical roles to induce transcription, recent reports suggest that lysine acetylation-mediated dissociation from negatively charged Glutamic acid-rich acidic domain-containing proteins has a significant impact on the gene expression. Detailed mechanisms of protein complex dissociation by acetylation may provide new ways to prevent or treat human diseases such as cancer, metabolic diseases and their complications.
Several modifications (phosphorylation, methylation, acetylation and others) of histones have been identified as important marks for controlling the human disease-related genes [1-3]. Lysine acetylation is a protein modification as binding sites for Bromodomain-containing 'reader' proteins (BRD) critical to induce gene expression [4]. However, a recent paper [5] reported that DNA damage-induced p21 expression is mediated by acetylation of p53 carboxyl terminal domain (CTD) and its dissociation from SET protein (Figure 1A) is involved in tumor regression in mouse xenograft models. They also showed that p53-interacting SET inhibits action of histone acetyltransferases (CBP/p300) and lowers p21 expression (Figure 1A). More importantly, the acidic domain (negatively charged Glutamic acid (E)-rich) in SET is critical to bind to lysine (positively charged) in p53CTD (Figure 1B). Therefore, unacetylated p53 tightly interacts with SET, however, acetylated p53 is dissociated from SET because the positive charges are neutralized by lysine acetylation (Figure 1B). The authors also identified known p53 binding proteins and many unknown candidate proteins which have acidic domain (negatively charged E-rich domain) as partners. Those proteins contain numerous factors related to DNA binding, transcriptional regulation, chromatin remodeling, suggesting that acetylation-mediated dissociation from acidic domain-containing proteins may be a key step to enhance the gene expression.
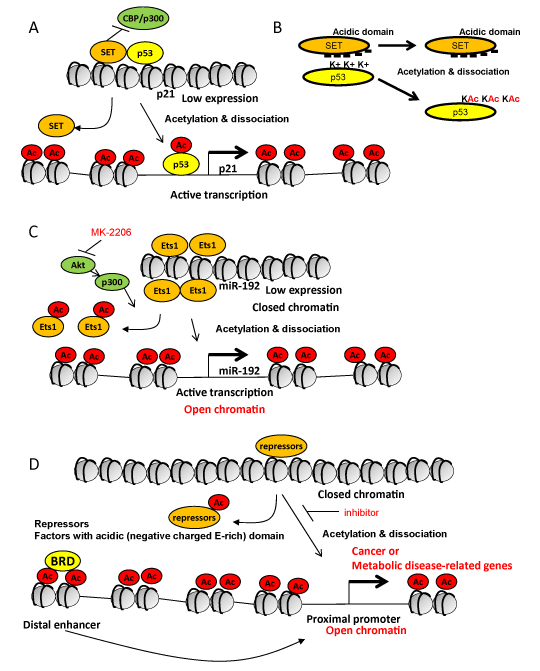
Figure 1. Lysine acetylation-mediated dissociation from acidic domain-containing proteins as a key step for the gene expression. (A) A model for DNA damage-induced p21 expression mediated by acetylation of p53 and its dissociation from SET protein in tumor regression in mouse xenograft models. p53-interacting SET also inhibits action of CBP/p300 and histone acetylation to inhibit p21 expression. Ac, acetylation. (B) Acidic domain (negatively charged Glutamic acid (E)-rich) in SET is critical to bind to lysine (positively charged) in p53. Unacetylated p53 tightly interacts with SET, however, acetylated p53 is dissociated from SET because the positive charges are neutralized by lysine acetylation (KAc). Ac, acetylation; -, negative charge; K+, positively charged lysine; KAc, acetylated lysine. (C) Lysine acetylation-mediated dissociation of Ets-1 and conformation changes in histones (from closed to open) in induction of miR-192 in the response to TGF-β related to fibrosis and hypertrophy in the pathogenesis of diabetic nephropathy. Lysine acetylation of Ets-1 (repressor of miR-192) and of histone H3 by p300 (activated by Akt) mediates dissociation of Ets-1 from the promoter region of miR-192 and conformation changes of histones (closed form to open form) in induction of miR-192. Akt inhibitor (MK-2206) prevents the signaling. (D) A simple and generalized model for active transcription mediated by lysine acetylation of histones and non-histone proteins through dissociation of protein complexes and conformation changes in nucleosomes or histones at the proximal promoter region. Combination of acetylation-mediated dissociation in the proximal promoter region and binding of bromodomain-containing 'reader' proteins (BRD) to lysine acetylation in the distal enhancer region may synergize to induce gene expression. Ac, acetylation; BRD, bromodomain-containing proteins.
On the other hand, microRNAs (miRs), such as miR-192, mediate the actions of transforming growth factor-β1 (TGF-β) related to fibrosis and hypertrophy in the pathogenesis of diabetic kidney diseases [3, 6]. In the process of induction of miR-192, lysine acetylation of the transcription factor Ets-1 (repressor of miR-192) and of histone H3 by the acetyltransferase p300 activated by the serine and threonine kinase, Akt, was also observed. Ets-1 is also known as a negative regulator of COL1A2 and acetylated Ets-1 (by p300) is dissociated from the COL1A2 promoter in human dermal fibroblast cells treated with TGF-β [7]. Therefore, lysine acetylation-mediated dissociation of repressor (Ets-1) and conformation changes of histones (closed form to open form) have critical roles in induction of miR-192 (Figure 1C). Interestingly, Akt inhibitor (MK-2206) was effective to decrease acetylation of Ets-1 and histones and prevent increase of miR-192 by TGF-β [6], suggesting a therapeutic potential through the mechanism.
Although there was no clear idea how acetylated histones or acetylated Ets-1 induce unwinding (closed to open) chromatin to induce the gene expression, many proteins related to chromatin, histones, nucleosome assembly have acidic domain which include negatively charged E-rich amino acid sequences [5]. Therefore, unacetylated Ets-1 may be tightly associated with those acidic domain-containing proteins in nucleosomes but acetylation may release Ets-1 nucleosomes. Also acetylated histones may be dissociated from those acidic domain-containing proteins in nucleosomes and change their forms from closed to open chromatin. A simple and generalized model is shown in Fig.1 D. Combination of acetylation-mediated dissociation in the proximal promoter region and binding of BRD to lysine acetylation in the distal enhancer region may synergize to induce gene expression [4] (Figure 1D).
Even in the other systems, lysine acetylation-mediated dissociation of protein complexes has been reported. Acetylation-dependent interaction between HMGB1 and SIRT1 is critical for LPS-induced macrophage lethality [8]. Lysine acetylation of adhesion molecules is also critical for dissociation from GAP junctions in dystrophic heart [9]. Acetylation of androgen receptor (AR) by Arrest-defective protein 1 (ARD1) induces its dissociation from Heat shock protein 90 complex to increase AR-regulated genes in the progression of prostate cancer [10]. Therefore, acetylation-mediated dissociation of protein complexes may be more broad or general mechanisms.
Since the genes regulated by histone acetylation are critical for cancer development, diabetes and diabetic complications, understanding of the mechanisms of controlling the protein interaction through acetylation would provide new therapeutic targets for those human diseases. In fact, because Bromodomain-containing proteins have critical roles to induce transcription through histone acetylation, some Bromodomain inhibitors are on the clinical trials for treatment of atherosclerosis, type 2 diabetes, and cancers [11]. However, the process of protein complex dissociation by acetylation may also be another possible target to control disease-related genes. Controlling acetylation-mediated dissociation may provide more unexpected ways to prevent or treat human diseases such as cancer, metabolic diseases and their complications.
References
- Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12(1): p. 7‐18. [Crossref]
- Zentner GE, Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 20: 259-266. [Crossref]
- Kato, M. and R. Natarajan, Diabetic nephropathy[mdash]emerging epigenetic mechanisms. Nat Rev Nephrol, 2014. 10(9): p. 517‐530. [Crossref]
- Marmorstein R, Zhou MM (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6(7): a018762. [Crossref]
- Wang D, Kon N, Lasso G, Jiang L, Leng W, et al. (2016) Acetylation‐regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538(7623): 118‐122. [Crossref]
- Kato M, Dang V, Wang M, Park JT, Deshpande S, et al. (2013) TGF‐beta induces acetylation of chromatin and of Ets‐1 to alleviate repression of miR‐192 in diabetic nephropathy. Sci Signal 6(278): ra43. [Crossref]
- Czuwara-Ladykowska J, Sementchenko VI, Watson DK, Trojanowska M (2002) Ets1 is an effector of the transforming growth factor beta (TGF‐ beta ) signaling pathway and an antagonist of the profibrotic effects of TGF‐beta. J Biol Chem 277(23): 20399‐408. [Crossref]
- Hwang JS, Choi HS, Ham SA, Yoo T, Lee WJ, et al. (2015) Deacetylation‐mediated interaction of SIRT1‐HMGB1 improves survival in a mouse model of endotoxemia. Scientific Reports, 2015. 5: p. 15971. [Crossref]
- Colussi C, Rosati J, Straino S, Spallotta F, Berni R, et al. (2011) Nε‐lysine acetylation determines dissociation from GAP junctions and lateralization of connexin 43 in normal and dystrophic heart. Proceedings of the National Academy of Sciences 108(7): 2795‐2800. [Crossref]
- DePaolo JS, Wang Z, Guo J, Zhang G, Qian C, et al., (2016) Acetylation of androgen receptor by ARD1 promotes dissociation from HSP90 complex and prostate tumorigenesis. Oncotarget 7(44): 71417‐71428. [Crossref]
- Filippakopoulos P, Knapp S (2014) Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13(5): 337‐56. [Crossref]

