Abstract
Triple negative breast cancer (TNBC) is a heterogeneous disease and has a higher rate of recurrence and distant metastasis. African-American (AA) women have a higher frequency of BRCA1 mutations and TNBC compared to other populations. Basal-like tumors have a higher rate of brain, lung and distant nodal metastasis more than other TNBC subtypes, contributing to higher mortality rate. Our previous work suggested Ubc9, a SUMO E2-conjugating enzyme to induce proliferation and migration of BRCA1-incompetent TNBC cells and TNBC cell lines established from the pleural effusion metastasis of a woman with TNBC. To understand the downstream signaling axis involved in distant metastasis we have used clinically relevant BRCA1 mutant and lung metastatic TNBC cell lines and our results show deregulated expression of caveolin-1, VEGF and SIRT1 in these cells compared to normal mammary epithelial cells by immunofluorescence analysis. We observed SIRT1 to be induced by wild type BRCA1a and BRCA1a I26A mutant unlike the disease associated Ubc9 binding mutants in TNBC cells. Knock down of Ubc9 induced SIRT1 expression in TNBC and ER-α expression in breast cancer cells. This is the first report demonstrating a role for Ubc9 in repressing both SIRT1 and ER-α expression in BRCA1 associated TNBC cells. It also suggests that the BARD-dependent E3 Ubiquitin ligase and HR (homologous recombination) activity of BRCA1 may not be required for inducing SIRT1 expression. Our results suggest for the first time that in BRCA1 mutant TNBC Ubc9-mediated induction of VEGF, inhibition of caveolin-1, SIRT1 and ER-α expression as a novel molecular mechanism underlying TNBC EMT (epithelial mesenchymal transition) leading to lung metastasis with pleural effusion. Drugs that target Ubc9 to both induce SIRT1 and ER-α or using SIRT1 agonists in combination with chemotherapy can be used as a promising targeted therapeutic approach for treating basal-like metastatic BRCA1-linked TNBC thus reducing the mortality in patients with TNBC.
Key words
BRCA1, Ubc9, Triple negative breast cancers, Cav-1, VEGF, SIRT1, ER-α, Lung metastasis, pleural effusion
Abbreviations
TNBC: triple negative breast cancers; ER: estrogen receptor; AA: african american; Cav-1: Caveolin1; VEGF: vascular endothelial growth factor; EMT: epithelial mesenchymal transition; HGSOC: high grade serous ovarian cancer; SIRT1: Sirtuin1
Introduction
Breast cancer is the most frequently diagnosed cancer in females in America, and it is the second leading cause of cancer deaths in America with 40,610 women expected to die from breast cancer in 2017 [1]. The breast cancer five-year survival rate drops dramatically to only 26.3% if distant metastasis occurs [2]. Triple Negative Breast Cancer (TNBC) is a heterogeneous disease that accounts for 10-20% of all breast cancers worldwide [3]. TNBC is notable for being an aggressive cancer with higher mean tumor size, higher grade tumors, higher rates of node positivity, high likelihood of recurrence, distinct metastasis patterns and poorer survival compared to other breast cancers [4]. TNBCs do not express estrogen receptor, (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) [4]. Because they do not express these receptors, TNBC’s lack targeted therapies that other breast cancers, such as ER+ breast cancer, have [4]. The Lehmann’s group classified TNBC into six molecular subtypes based on their gene expression profiles: Basal-Like-1 (BL1), Basal-Like-2 (BL2), Luminal Androgen Receptor (LAR), Mesenchymal Stem-Like (MSL), Mesenchymal (M), and Immunomodulatory (IM) [3-5]. Most TNBC with BRCA1 mutations fall into the basal-like subtypes (BL1 and BL2), which have high rates of recurrence and distant metastasis, especially to the lungs and brain, contributing to higher mortality. African-American and Hispanic women are disproportionally affected by TNBC Basal-Like cancers with BRCA1 mutations, meaning they also have worse outcomes than women of other ethnicities. BL1 and BL2 subtypes with BRCA1 mutation are usually treated with platinum-based chemotherapy like cisplatin [5], but recurrence and metastasis are still an issue [6,7]. There is thus a critical need to obtain druggable targets so as to develop targeted therapy for TNBC. BRCA1 is a tumor suppressor gene located on the long arm of chromosome 17 that is either mutated, expressed at low levels, or localized abnormally in TNBC [4,8]. We have cloned two major splice variants of BRCA1, namely BRCA1a/p110 and BRCA1b/p100 [9,10], both are expressed at lower levels in breast tumors as opposed to normal cells [11-14]. We have found BRCA1a protein to induce apoptosis and inhibit in vivo tumor growth of TNBC cells [15,16]. BRCA1 proteins contain several functional domains, an N-terminal RING finger domain that interacts with multiple proteins and two-BRCA1 C-terminal domains involved in transcriptional activation. BRCA1, BRCA1a and BRCA1b proteins are nuclear-cytoplasmic shuttling proteins that are also localized in the mitochondria [9,14,17,18]. The action of nuclear localization signals (NLS) and nuclear export signals (NES) located in the RING domain that via association with BARD1 also regulate BRCA1 nuclear transport [19]. The BRCA1 delta isoform, which lacks NLS, also enters the nucleus via the BARD1 import pathway [19]. The RING domain of BRCA1, in complex with BARD1, mediates an E3 Ubiquitin ligase activity on ER-α in-vitro [20, 21]. Using an Ubiquitin ligase-deficient BRCA1 I26A mutant some groups have found that the Ubiquitin ligase activity is expendable for both, genomic stability and homology-directed repair of double-strand DNA breaks, however the Ubiquitin ligase activity is essential for repression of ER-α activity [22,23]. The SUMO pathway was shown to be involved in BRCA1 response to DNA damage and transcriptional repression [24,25]. We have shown the amino-terminal domain of BRCA1, BRCA1a and BRCA1b proteins to bind to SUMO-E2-conjugating enzyme Ubc9 and activate ER-α activity by promoting its degradation in vivo [26]. This work suggested for the first time a new biochemical activity of BRCA1 as a putative SUMO-1 and Ubc9-dependent E3 Ubiquitin ligase for ER-α [26]. The cancer-predisposing mutation in BRCA1 (C61G), disrupted the ability to regulate Ubc9-mediated estrogen-induced ER-α transcriptional activity in breast cancer cells [26] suggesting for the first time a molecular mechanism for TNBC development due to BRCA1 mutation. Both BRCA1/BRCA1a K109R and C61G mutants that are localized mostly in the cytoplasm, fail to inhibit the growth of TNBC cells [27]. Ubc9 plays a critical role in breast cancer cell migration, tumor progression and resistance to chemotherapy [28-33]. Ubc9 expression has also been found to correlate with poor clinical outcome in Nigerian black women with breast cancer [34]. Our recent results suggests knockdown of Ubc9 in BRCA1 mutant TNBC and HGSOC cells to inhibit proliferation and migration suggesting a role in epithelial to mesenchymal transition (EMT) of TNBC [35]. EMT is a biological phenomenon wherein epithelial cells gain the ability to migrate to distal sites, playing a key role in cancer metastasis. Similarly we have also observed elevated expression of Ubc9 in HGSOC with BRCA1 mutation which resulted in loss of caveolin-1 and induction of VEGF suggesting a pathway linking BRCA1 mutation to HGSOC with peritoneal permeability and ascites formation [36]. Sirtuin 1 (SIRT1), a nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase linked to longevity, metabolism, stress responses, genomic stability, energy homeostasis and ER-α repression [37, 38]. Dr. Deng‘s group demonstrated SIRT1 expression to be reduced in BRCA1-associated breast cancers and BRCA1 was shown to bind to the SIRT1 promoter and increase its expression which inhibits Survivin levels [39]. These studies clearly suggest BRCA1 to play a tumor suppressor role through regulation of SIRT1 expression [40]. Recently hypoxia, a promoter of EMT was shown to repress SIRT1 expression in a SUMOylation-dependent manner [41]. This data suggests SIRT1 to function as a tumor suppressor in breast, ovarian and lung cancer cells [37-41]. In this study we have identified the underlying molecular mechanism of BRCA1 –associated TNBC metastasis to the lung so as to discover novel biomarkers for developing targeted therapies for TNBC. We have found reduced levels of caveolin-1 and SIRT1 and elevated expression of VEGF in both BRCA1 mutant TNBC cells as well as TNBC cells established from a pleural effusion lung metastasis. Ubc9 knockdown induced both SIRT1 and ER-α expression in breast cancer cells. Furthermore, these results suggested that Ubc9 binding but not BARD-dependent E3 Ubiquitin ligase activity and HR activity of BRCA1 is required for SIRT1 induction. Both BRCA1 and SIRT1 induced ER-α expression in BRCA1 mutant TNBC cells. Our data provides for the first time a novel mechanism as to how BRCA1 by tethering Ubc9 a tumor promoter induces SIRT1, caveolin-1 and ER--α expression inhibiting TNBC metastasis to lung.
Materials and methods
Cell culture
MCF10A, CAL51, T47D, HCC1937, and HCC1937 BRCA1 cells were obtained from American Type Culture Collection (Rockville, MD, USA), CAL51 cells were obtained from CD Biosciences Inc. and cultivated as described previously [26-28]. MCF10A were grown in 50% RPMI and 50% MEGM (CC-3150, Lonza), supplemented with 3% fetal bovine serum. CAL51 were grown in DMEM medium with 10% FBS, 0.6µg/ml Insulin, 5X103 µg/ml transferrin and 1% PS. T47D were grown in DMEM medium with 10% FBS, 0.2 units/ml Insulin and 1% PS. HCC1937 cells were grown in RPMI 1640 medium with 10% FBS and 1% PS.
Expression plasmids
The BRCA1/1a expression plasmids BRCA1a, BRCA1a Mut#1 (K109R), BRCA1a Mut#4 (C61G), BRCA1a Mut#9 (I26A), EGFP-AS Ubc9, Flag SIRT1 were cloned as described previously [27, 35].
Western blot analysis
CAL51 cells were seeded into 10 cm Petri-dishes with a density of 2 x 106. Cells were transfected with EGFP or EGFP-Antisense Ubc9 plasmids. After 48 hours of transfection, cells were collected and the cell pellets were lysed in RIPA lysis buffer (50 mm Tris-HCl, (pH7.8) 150 mm NaCl, 5 mm EDTA, 0.5% Triton X-100, 0.5% NP40, 0.1% sodium deoxycholate) and the proteins were separated on 4-20% gradient SDS-PAGE and transferred to nitrocellulose membrane. The primary antibodies for Ubc9 (Santa Cruz, N-15, 1/150) and SIRT1 (Santa Cruz, H-300, 1/150) and β-Actin (Santa Cruz, C4, 1/1000) were used to probe the proteins of interest. The protein bands were visualized using HRP labeled secondary antibody to mouse or rabbit or goat and developing solution (GE Healthcare) by Image Reader LAS-3000 (FUJIFILM). The values were standardized with the internal control β-Actin.
Antibodies and reagents
The antibodies used in this study were primary polyclonal Rabbit anti-caveolin-1 antibody (Santa Cruz biotechnology), primary polyclonal Rabbit anti-VEGF antibody (Santa Cruz Biotechnology), primary polyclonal Rabbit anti-SIRT1 antibody (Santa Cruz Biotechnology), primary polyclonal Goat anti-Ubc9 N-15 antibody (Santa Cruz Biotechnology), primary monoclonal mouse anti- β-Actin antibody (Santa Cruz Biotechnology), primary monoclonal mouse anti- ER-α antibody (Santa Cruz Biotechnology).
Immunofluorescence analysis
MCF10A, HCC1937, HCC1937 BRCA1 and CAL51 cells were cultured alone or transfected with pCDNA3, BRCA1a, BRCA1a Mut#1, BRCA1a Mut#4, BRCA1a Mut#9 or EGFP-Antisense Ubc9 for 24 hours in six-well plates onto glass coverslips overnight. The cells were washed and fixed in icy methanol for 5 minute, and blocked using 10% BSA for 60 min, followed by primary polyclonal Rabbit anti-caveolin-1 antibody 1:150, primary polyclonal anti-SIRT1 antibody (Santa Cruz),1:150 diluted in 1.5% BSA made in PBS at 25°C for 1hr and Alexa 488 goat anti-Rabbit/Alexa 568 goat anti-mouse (Molecular Probes) diluted in 1.5% BSA/PBS for 50 min and stained (Hoechst 33258, Pentahydrate, Life technologies). The cover slips were mounted with Vectashield mounting medium for fluorescence (H-1000 from Vector). The stained cells were examined by LSM 700 Confocal Microscope, equipped with 63X oil Ph immersion objectives. Composite filter cubes were used for the 488-405 as described previously [27].
siRNA transfection and western blot analysis
T47D cells were maintained in DMEM medium, supplemented with 10% fetal bovine serum. Before transfection, cells were digested with 0.25% of Trypsin-EDTA solution and seeded into 10 cm diameter petri dishes with a density of 1.5 x 105/well. Twenty-four hours later, dilute Ubc9 siRNA and control siRNA (Qiagen) in the culture medium without serum (final concentration is 50 nM), HiPerFect Transfection Reagent (Qiagen) was added to the diluted siRNA and mixed by vortexing. The mixture was incubated for 5–10 min at room temperature (15–25°C) to allow the formation of transfection complexes. The complexes were added drop-wise onto the cells and gently swirled to ensure uniform distribution of the transfection complexes. The cells were incubated for 48 hours and subjected to western blot analysis using Ubc9 and ERα antibodies as described previously [26,27].
Results
Caveolin-1 expression is reduced in BRCA1 mutant TNBC cells
Wild Type BRCA1 protein stimulates caveolin-1 expression by binding to the caveolin-1 promoter and caveolin-1 also induces BRCA1 protein expression [42,43]. To examine whether this occurs in a physiological relevant BRCA1 mutant cell line in vivo, we have studied the expression of caveolin-1 in normal human mammary epithelial cells MCF10A and a Basal-Like TNBC cell line HCC1937 obtained from a patient with germ line BRCA1 mutation using immunofluorescence analysis. Our results show reduced levels of expression of caveolin-1 in HCC1937 but not in MCF10A cells (Figure 1). These results suggest BRCA1 protein to induce caveolin-1 expression in normal mammary epithelial cells and BRCA1 dysfunction results in down regulation of caveolin-1 in BRCA1 mutant TNBC cells. These results support our previous observation in BRCA1 mutant HGSOC cells and what was observed by others [35,43].
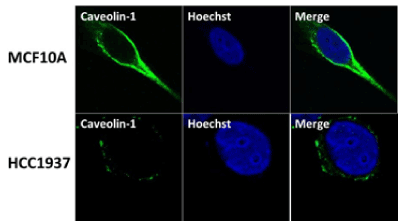
Figure 1. Caveolin-1 is expressed at reduced levels in BRCA1 mutant TNBC cells HCC1937 compared to normal mammary epithelial cells MCF10A using immunofluorescence analysis. MCF10A and HCC1937 cells were cultured in six-well plates onto glass cover slips overnight. cells were washed and fixed in icy methanol for 5 minute, and blocked using 10% BSA for 60 min, followed by primary polyclonal Rabbit anti-caveolin-1 antibody (Santa Cruz) 1:150 diluted in 1.5% BSA/PBS at 25°C for 1hr and Alexa 488 goat anti-Rabbit (Molecular Probes) 1:100 diluted in 1.5% BSA/PBS for 50 min, in combination with Hoechst staining (Life technologies). The cover slips were mounted with Vectashield mounting medium for fluorescence analysis. The stained cells were examined by LSM 700 Confocal Microscope, equipped with 63X oil lenses.
VEGF is expressed at elevated levels in BRCA1 mutant TNBC cells compared to normal mammary epithelial cells
BRCA1 was shown to inhibit VEGF expression and protein secretion through the estrogen signaling pathway in breast cancer cells [44] and increased VEGF levels were found in tumor tissue of BRCA1/2 carriers [45]. To examine whether the same scenario is true in BRCA1 mutant TNBC cells we have studied the expression of VEGF using immunofluorescence analysis. Our results show elevated levels of VEGF in HCC1937 cells compared to normal mammary epithelial cells MCF10A (Figure 2). These results indicate a role for BRCA1 in regulating angiogenesis as previously observed by us in BRCA1 mutant HGSOC cells [35,36] and by others in BRCA1/2 –related breast tumors [45].
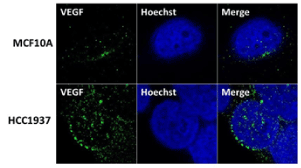
Figure 2. VEGF is expressed at elevated levels in BRCA1 mutant TNBC cells HCC1937 compared to normal mammary epithelial cells MCF10A. VEGF proteins were detected by immunostaining using VEGF antibodies. MCF10A, HCC1937 cells were cultured in six-well plates onto glass cover slips overnight. cells were washed and fixed in icy methanol for 5 minute, and blocked using 10% BSA for 60 min, followed by primary Polyclonal Rabbit anti-VEGF antibody (Santa Cruz) 1:150 diluted in 1.5% BSA/PBS at 25°C 1hr and Alexa 488 goat anti-Rabbit antibody (Molecular Probes) 1:100 diluted in 1.5% BSA/PBS for 50 min, in combination with staining with Hoechst (life technologies). The cover slips were mounted with Vectashield mounting medium for fluorescence (Vector). The stained cells were examined by LSM 700 Confocal Microscope, equipped with 63X oil immersion objectives.
BRCA1 induces expression of endogenous SIRT1 in TNBC cells
BRCA1 -associated breast cancers have lower levels of SIRT1 than their normal controls [40]. SIRT1 expression was found to be down-regulated by hypoxia which promotes EMT and lung cancer metastasis [41]. To study whether low levels of SIRT1 expression is responsible for BRCA1 –associated Basal-Like TNBC tumors to have higher rate of metastasis to the lung and brain we studied the expression of SIRT1 in BRCA1 mutant TNBC cells HCC1937 and a sporadic TNBC cell line CAL51 established from the pleural effusion metastasis of a 45-year –old woman with progressive breast adenocarcinoma (after radio-chemotherapy and surgery) using immunofluorescence analysis. Our results show low level of expression of SIRT1 in both the TNBC cell lines compared to BRCA1 reconstituted HCC1937 cells (see Figure 3). These results suggest a role for SIRT1 in metastasis by regulating EMT as shown by others in lung cancers [41].
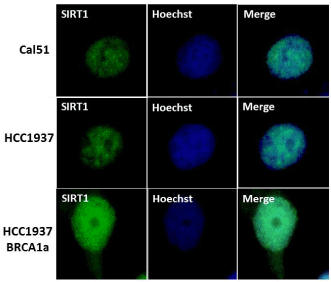
Figure 3. CAL51 and BRCA1 mutant TNBC HCC1937 cell lines were found to express lower levels of SIRT1 compared to BRCA1 reconstituted HCC1937 cells using immunofluorescence analysis. Cal51, HCC1937, HCC1937+BRCA1 Cells were cultured in six-well plates onto glass cover slips overnight. cells were washed and fixed in icy methanol for 5 minute, and blocked using 10% BSA for 60 min, followed by primary monoclonal anti-Sirt1 antibody (Santa Cruz) 1:150 diluted in 1.5% BSA/PBS at 25°C 1hr and Alexa 488 goat anti-Mouse antibody (Molecular Probes) diluted in 1.5% BSA/PBS for 50 min, in combination with staining with Hoechst (Life technologies).The cover slips were mounted with Vectashield mounting medium for fluorescence (Cat# H-1000 from Vector). The stained cells were examined by LSM 700 Confocal Microscope, equipped with 63X oil Ph immersion objectives.
BRCA1a pathogenic mutants fail to induce SIRT1 expression unlike wild type BRCA1a and E3 Ubiquitin ligase deficient BRCA1a mutant in TNBC cells established from lung pleural effusion
Wildtype BRCA1/1a proteins were shown by us to bind to SUMO conjugating enzyme Ubc9 and inhibit the tumor growth of TNBC cells unlike the disease associated pathogenic mutants [27]. To test whether the various BRCA1a mutants (BRCA1a Mut#1, BRCA1a Mut#4, and BRCA1a Mut #9) can induce SIRT1 similar to wild type BRCA1/BRCA1a proteins, we have studied the expression of SIRT1 in CAL51 cells using immunofluorescence analysis. Our results show BRCA1a Mut#1 and Mut#4 to express low levels of SIRT1 compared to wild type BRCA1a and BRCA1a Mut#9 (Figure 4). These results suggest that Ubc9 binding may be required for inducing SIRT1 expression by BRCA1/1a in TNBC cells. BRCA1/1a proteins by tethering Ubc9 may prevent it from inhibiting SIRT1 activity in a SUMOylation dependent manner [41]. These results also indicate that the E3 Ubiquitin ligase activity and HR activity is not required for inducing SIRT1 expression.
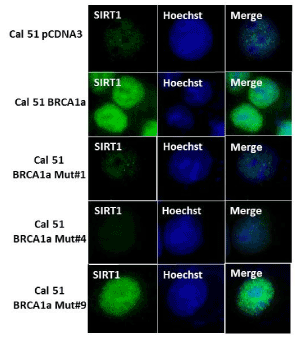
Figure 4. Wild type BRCA1a and BRCA1a I26A mutant but not the C61G and K109R mutants induce SIRT1 expression in CAL51 TNBC cells as detected by immunofluorescence analysis.CAL51 cells were seeded into six-well plates and transfected with pcDNA3 or pcDNA3 BRCA1a or BRCA1a Mut#1, BRCA1a Mut#4 or BRCA1a Mut#9 as described in Figure 3. The stained cells were examined by LSM 700 Confocal Microscope, equipped with 63X oil Ph immersion objectives.
Knockdown of Ubc9 restores SIRT1 expression in TNBC cells established from lung pleural effusion
BRCA1 has been shown to bind to the SIRT1 promoter and induce its expression in HCC1937 cells [39]. We have previously used wound healing assay to show that knockdown of Ubc9 in HCC1937 and CAL51 cells inhibits migration of these cells [35]. To test whether knockdown of Ubc9 can restore SIRT1 expression in CAL51 TNBC cells which express high levels of Ubc9, we suppressed Ubc9 expression in CAL51 cells by using antisense Ubc9 and measured SIRT1 expression by western blot analysis. As shown in Figure 5A and B, Ubc9 knockdown resulted in increased expression of SIRT1 compared to control vector transfected cells. Together, these results indicate that Ubc9 is essential for TNBC cell migration and downregulation of Ubc9 may facilitate an inhibitory effect of SIRT1 on TNBC cell migration.
Ubc9 knockdown induces ER-α expression in ER- α positive breast cancer cells
BRCA1 proteins have been found by us to induce ER-α expression by tethering Ubc9 and BRCA1 pathogenic mutants do not bind Ubc9, form colonies in soft agar and are transforming [27]. Based on these findings we developed a BRCA1-function based assay [26] and a patent has been issued for this work (Number 8, 372,580). If Ubc9 is responsible for repressing ER-α expression then knockdown of Ubc9 in breast cancer cells should reactivate ER-α. To test this we knockdown Ubc9 expression in T47D breast cancer cells using Ubc9 siRNA and studied ER-α protein expression by western blot analysis. As shown in Figure 5C, Ubc9 downregulation using Ubc9 siRNA resulted in increased expression of ER-α protein compared to control siRNA transfected T47D cells. These results clearly indicate that Ubc9 inhibits ER-α activity in breast cancer cells. Ubc9 was shown to inhibit ER-α activity in the presence of SUMO-1 and this was thought to be due to extensive SUMOylation of ER-α and/or its cofactors which could result in this repression [46]. This could be a scenario one sees in TNBC where there is mutation in the BRCA1 gene and ER-α is not expressed [26].
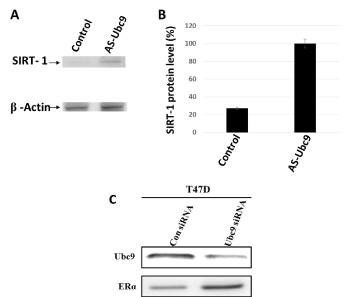
Figure 5. Ubc9 knockdown increases SIRT1 and ER- α expression in CAL51 cells established from the pleural effusion metastasis of a patient with TNBC and T47D ER- α positive breast cancer cells. (A). CAL51 cells were seeded into 10cm Petri dishes overnight. Following the transfection with AS Ubc9 plasmid or control plasmid for 48hrs the cells were collected and lysed in RIPA buffer. The cell lysates were probed with primary antibodies either Ubc9 (Santa Cruz N15, 1/100) or β Actin (Santa Cruz C4, 1/2000 or 10009 (Santa Cruz, 1/100). The proteins were visualized using HRP labeled secondary antibody to goat (Ubc9) or mouse (β Actin) and developing solution (GE Healthcare) through Image Reader LAS-3000 (FUJIFILM). (B).The stoichiometry of SIRT1 protein levels shown in A. The signal of SIRT1 protein band was quantified using software Multiguage. The values were standardized with the internal control β Actin, CAL51 is defined as 1. (C). T47D cells were seeded into the Petri dishes of 10 cm diameter 24 hours before transfection. HiPerFect Transfection Reagent (Qiagen) was used to mediate either Ubc9 or control siRNA transfection. Following 48 hours of siRNA transfection, the cells were lysed and subjected to Western blot analysis using Ubc9 and ERα antibodies.
BRCA1 and SIRT1 induce ER-α expression in BRCA1 mutant TNBC cells
SIRT1 regulates ER-α expression and SIRT1 knockout reduces ER-α protein in mouse mammary gland [47]. BRCA1 was shown to inhibit the ligand dependent and independent transcription activity of ER-α [38, 48]. We have previously shown that BRCA1 disease associated mutants were defective in their ability to bind Ubc9 causing ER-α repression and thus promoting tumorigenesis [26,27,35]. We therefore tested whether SIRT1 and BRCA1 can activate ER-α expression in BRCA1 mutant TNBC cell line HCC1937. As shown in figure 6A and B SIRT1 and BRCA1 induced ER-α expression in HCC1937 cells. Taken together the data indicates that both SIRT1 and BRCA1 are able to independently induce ER-α expression in BRCA1 mutant TNBC cells.
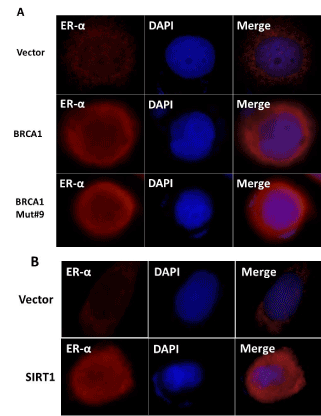
Figure 6. Wild type BRCA1, BRCA1 Mut#9 (I26A) and SIRT1 induce expression of ER-α in HCC1937 TNBC cells as seen by immunofluorescence analysis. (A) HCC1937 cells were seeded into six-well plates and transfected with pCDNA3, BRCA1, BRCA1 I26A, (B) CMV Flag and Flag SIRT1 as described in Figure 3. The nuclei were visualized with DNA staining dye Hoechst 33258. Cells were fixed and probed with ER-α (Santa Cruz, 1/250) followed by Alexa Fluor 568 labeled secondary antibody (Invitrogen, 1/200) staining. The images were taken using LSM 700 confocal microscope (100X, oil Ph immersion objectives).
Discussion and conclusions
Metastatic TNBC is a very aggressive type of breast cancer and has the highest mortality rate in comparison to other breast cancers. BRCA1 mutation carriers belong to the TNBC Basal-like subtype and lack effective targeted therapies [3,4]. Hence treatments are limited to surgery, radiotherapy and chemotherapy [5]. There is thus an urgent need to develop novel targeted therapy for metastatic TNBC. Tumor VEGF expression is significantly higher in TNBC compared to non TNBC [49]. In this study we have found high levels of VEGF, reduced levels of caveolin-1, SIRT1 in highly migratory BRCA1 mutant TNBC cells and TNBC cells established from a pleural effusion metastasis of a women with TNBC (after radio-, chemotherapy and surgery) compared to normal mammary epithelial cells. This agrees with our earlier work showing reduced expression of caveolin-1 and higher levels of VEGF in BRCA1 mutant HGSOC cells suggesting a role for VEGF in cancer metastasis (35). Sun et al. [41] have found activation of SIRT1 to result in inhibition of migration of lung cancer cells in vitro and lung metastasis in vivo. Our previous published results showed SUMOylation enzyme Ubc9 to play a key role in migration of BRCA1 mutant TNBC and HGSOC cells [35] which agrees with our findings. We observed BRCA1/BRCA1a proteins but not the disease associated pathogenic BRCA1 mutants, which do not bind Ubc9 to induce endogenous SIRT1 expression in TNBC cells. Another BRCA1 mutant I26A which lacks both E3 Ubiquitin ligase activity and HR activity was found to induce SIRT1 expression in TNBC cells. Knockdown of Ubc9 in TNBC cells resulted in expression of SIRT1 in these cells suggesting inhibition of SIRT1 activity by SUMO E3 conjugase Ubc9. These results suggest a role for Ubc9 in promoting EMT of TNBC cells causing lung metastasis which is consistent with a recent study showing a SUMOylation-dependent pathway to regulate SIRT1 transcription and lung cancer metastasis [41]. We have identified SIRT1 as a new target that is downregulated in BRCA1 mutant TNBC cells. BRCA1 was shown previously to induce SIRT1 expression by binding to its promoter [39]. Our results suggest yet another novel mechanism whereby BRCA1 by binding Ubc9 activates SIRT1 expression. BRCA1 and SIRT1 both also repress ER- α -mediated transcriptional activity and proliferation in estrogen responsive breast cancer cells [38]. Knock down of Ubc9 in ER- α positive breast cancer cells T47D resulted in induction of ER- α expression, once again suggesting a role for SUMOylation in repressing ER-α activity in breast cancer cells. To our knowledge this is the only mechanism that can explain how BRCA1 mutation can result in TNBC [26]. Based on these findings a BRCA1 function-based cellular assays patent has been issued on this work (patent number 8, 372,580). These studies suggest a novel Ubc9-caveolin-1-VEGF-SIRT1- ER- α axis that when perturbed due to BRCA1 dysfunction results in TNBC EMT and distant lung metastasis (Figure 7). Our findings along with that of others indicate BRCA1 and it downstream target SIRT1 to play a major role in inhibiting metastasis of TNBC. Future studies will be geared towards targeting Ubc9 to both induce SIRT1 and ER- α expression or using SIRT1 agonists in combination with chemotherapy to provide a novel next- generation targeted therapy to treat Basal-like BRCA1-associated TNBC metastasis to lung.
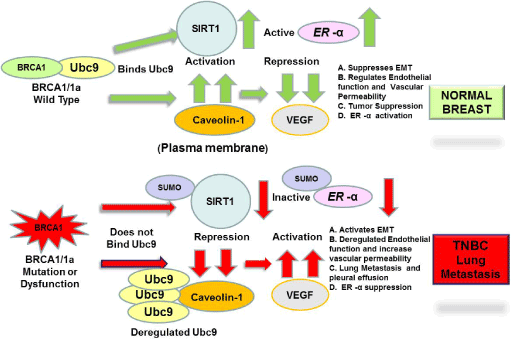
Figure 7.
Acknowledgements
We thank all the members of Drs. Rao and Reddy labs for their help. We thank Mr. Abramson, research media services and Ms Wimes for editorial assistance and RCMI core facilities at Morehouse School of Medicine, for their assistance. This work was supported in part by Georgia Cancer Coalition Distinguished Cancer Scholar award, NIH-NCRR-RCMI grant G-12-RR003034, U54 RR02613, 5P20RR11104 and NIHMD research endowment grant 2S21MD000101 and U54 CA118638 to V.N.R. V.N.R’s lab was also supported in part by private funds from the VOYA foundation and It’s the Journey Inc.
Conflict of interest
None.
References
- American Cancer Society. Cancer Facts & Figures 2017. Atlanta: American Cancer Society.
- SEER Cancer Statistics Factsheets: Female Breast Cancer. National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/statfacts/html/breast.html (http://seer.cancer.gov/statfacts/html/breast.html)
- Aysola K, Desai A, Welch C, Xu J, Qin Y, et al. (2013)Triple Negative Breast Cancer- An overview. Hereditary Genetics, Special Issue: Hereditary Disorders. [Crossref]
- Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA (2014) New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clin Cancer Res 20: 782-790. [Crossref]
- Le Du P, Eckhardt BL, Lim B, Litton JK, Moulder S, et al. (2015) Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype? Oncotarget 6(15): 12890-908. [Crossref]
- Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, et al. (2010) Metastatic behavior of breast cancer subtypes. J Clin Oncol 28: 3271-3277. [Crossref]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66-71. [Crossref]
- Wilson C, Ramos L, Villasenor M, Anders K, Press M, et al. (1999) Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nature Genetics 21:236-240. [Crossref]
- Wang H, Shao N, Ding OM, Cui J, Reddy ESP, Rao VN (1997) BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phophoprotein that associate with E2F, cyclin and cyclin dependent kinases. Oncogene 15: 143-157.
- Chai YL, Cui J, Chipitsyna G, Liao B, Lui S, Aysola K, et al. (2001) C-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b mediated growth suppression in breast cancer cells. Oncogene 20: 1357-1367. [Crossref]
- Wilson CA, Payton MN, Elliott ES, Buaas FW, Cajulis EE, et al. (1997) Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta 11b. Oncogene 14: 1-16. [Crossref]
- Lu M, Conzen SD, Cole CN, Arrick B (1996) Characterizations of functional messenger RNA splice variants of BRCA1 expressed in non-malignant and tumor-derived breast cells. Cancer Res 56: 4578-4581. [Crossref]
- Orban TI, Olah E (2001) Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem Biophys Res Commun 280: 32-38. [Crossref]
- Maniccia AW, Lewis C, Begum N, Xu J, Cui J, et al. (2009) Mitochondrial localization, ELK-1 transcriptional regulation and Growth inhibitory functions of BRCA, BRCA1a and BRCA1b proteins. J Cell Physiol 219: 634-641. [Crossref]
- Shao N, Chai YL, Shyam E, Reddy P, Rao VN (1996) Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene 13: 1-7. [Crossref]
- Chai Y, Shao N, Lee L, Reddy V, Gabriela O, et al. (2007) BRCA1a has antitumor activity in Triple-negative breast and ovarian and prostate cancer cells. Oncogene 26: 6031-6037. [Crossref]
- Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, et al. (2005) Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell 16: 997-1010. [Crossref]
- Rosen EM, Fan S, Ma Y (2006) BRCA1 regulation of transcription. Cancer Lett 236: 175-185. [Crossref]
- Fabbro M, Rodriguez JA, Baer R, Henderson BR (2002) BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem 277: 21315-21324. [Crossref]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, et al. (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537-14540. [Crossref]
- Eakin CM, Maccoss MJ, Finney GL, Klevit RE (2007) Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci U S A 104: 5794-5799. [Crossref]
- Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, et al. (2008) E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci U S A 105: 20876-20881. [Crossref]
- Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, et al. (2010) BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol 24: 76-90. [Crossref]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, et al. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886-890. [Crossref]
- Park MA, Seok YJ, Jeong G, Lee JS (2008) SUMO1 negatively regulates BRCA1-mediated transcription, via modulation of promoter occupancy. Nucleic Acids Res 36: 263-283. [Crossref]
- Xu J, Watkins T, Reddy A, Reddy ES, Rao VN. (2009) A novel mechanism whereby BRCA1/1a/1b fine tunes the dynamic complex interplay between SUMO-dependent/independent activities of Ubc9 on E2-induced ERalpha activation/repression and degradation in breast cancer cells. Int J Oncol 34: 939-949. [Crossref]
- Qin Y, Xu J, Aysola K, Begum N, Reddy V, et al. (2011) Ubc9 mediates nuclear localization and growth suppression of BRCA1 and BRCA1a proteins. J Cell Physiol 226: 3355-3367. [Crossref]
- Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, et al. (2009) Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int J Cancer 125: 596-602. [Crossref]
- Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD, Jr., Annunziata CM, Munshi NC. (2010) The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood 115: 2827-2834. [Crossref]
- Mo YY, Yu Y, Ee PL, Beck WT. (2004) Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res 64: 2793-2798. [Crossref]
- Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, et al. ( 2010) Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol 41: 1286-1298. [Crossref]
- He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, et al. (2015) Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One 10: e0123882. [Crossref]
- Chen SF, Gong C, Luo M, Yao HR, Zeng YJ, et al. (2011) Ubc9 expression predicts chemoresistance in breast cancer. Chin J Cancer 30: 638-644. [Crossref]
- Agboola AO, Musa AA, Ayoade BA, Banjo AA, Anunobi CC, et al. (2014) Clinicopathological and molecular significance of Sumolyation marker (ubiquitin conjugating enzyme 9 (UBC9)) expression in breast cancer of black women. Pathol Res Pract: 10-7. [Crossref]
- Xu J, Footman A, Qin Y, Aysola K, Black S, et al. (2016) BRCA1 Mutation Leads to Deregulated Ubc9 Levels which Triggers Proliferation and Migration of Patient-Derived High Grade Serous Ovarian Cancer and Triple Negative Breast Cancer Cells. Int J Chronic Dis Ther 2: 31-38. [Crossref]
- Desai A, Xy J, Aysola K, Akinbobuyi O, White M, et al. (2015) Molecular Mechanism linking BRCA1 Dysfunction to HGSEOC with peritoneal permeability and ascites. J Gynecol Res 1: 1-7. [Crossref]
- Revollo JR, Li X (2013) The ways and means that fine tune Sirt1 activity. Trends Biochem Sci 38: 160-167. [Crossref]
- Moore R, Faller D (2013) SIRT1 represses estrogen-signaling, ligand-independent ERa-mediated transcription, and cell proliferation in estrogen-responsive breast cells. J Endocrinol 216: 273-285.
- Wang RH, Zheng Y, Kim HS, Xu X, Cao L, et al. (2008) Interplay among BRCA, SIRT, and Survivin during BRCA1-associated tumorigenesis. Mol Cell 32: 11-20. [Crossref]
- Deng CX (2009) SIRT, is it a tumor promoter or tumor suppressor? Int J Biol Sci 5: 147-152. [Crossref]
- Sun L, Li H, Chen J, Dehennaut V, Zhao Y, et al. (2013) A SUMOylation-dependent pathway regulates SIRT1 transcription and lung cancer metastasis. J Natl Cancer Inst 105: 887-898. [Crossref]
- Glait Chen, Ravid Dana, Lee Sam W, Mordechai L, Werner Haim (2006) Caveolin-1 controls BRCA1 gene expression and cellular localization in human breast cancer cells. FEBS Letters, 580.
- Wang Y, Yu J, Zhan Q (2008) BRCA1 regulates caveolin-1 expression and inhibits cell invasiveness. Biochem Biophys Res Commun 370: 201-206. [Crossref]
- Kawai H, Li H, Chun P, Avraham S, Avraham HK (2002) Direct interaction between BRCA1 and the estrogen receptor regulates vascular endothelial growth factor (VEGF) transcription and secretion in breast cells. Oncogene 21: 7730-7739. [Crossref]
- Danza K, De Summa S, Pinto R, Pilato B, Palumbo O, et al. (2015) MiR-578 and miR-573 as potential players in BRCA-related breast cancer angiogenesis. Oncotarget 6: 471-483. [Crossref]
- Sentis S, Le Romancer M, Bianchin C, Rostan MC, Corbo L (2005) Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol 19: 2671-2684. [Crossref]
- Yao Y, Li H, Gu Y, Davidson NE, Zhou Q (2010) Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis 31: 382-387. [Crossref]
- Fan S, Wang J, Yuan R, Ma Y, Meng Q, et al. (1999) BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science 284: 1354-1356. [Crossref]







