The human respiratory syncytial virus (HHRSV) has a remarkable importance as a causative agent of respiratory tract infection in children. Preterm infants or those affected by some chronic conditions are especially compromised by HHRSV infection. Monoclonal antibodies against HRSV F protein have been used to prevent severe disease. This study evaluates a group of 58 newborns (median age 10 months) in use of Palivizumab® prophylaxis presenting any symptom of respiratory tract infection by tests currently available. Positive nasopharyngeal samples were then analyzed and viral load compared to a control group of non-risk outpatient’s children without prophylaxis.
The detection was 15.9% (11/69) with any of the assays, and increased with the use of molecular techniques (36.3% for qPCR). Out of 58 children in use of palivizumab four developed bronchilolitis.
There was no difference on viral load between children in prophylaxis with severe disease or mild symptoms and children without prophylaxis (p = 0.4).
According to this preliminary data monoclonal antibody mechanism of disease suppression seems not to be directly correlated to the amount of viral replication.
respiratory virus, molecular methods, palivizumab, HHRSV viral load
Human respiratory syncytial virus (HHRSV) is the most important causative agent of lower respiratory tract infection in children and infants [1,2].
Preterm infants or those affected by some chronic conditions are especially compromised by HHRSV infection usually accounting for long periods of hospitalization and high rates of mortality [3]. Prevention from infection is possible through the use of drugs that can provide passive immunity against HHRSV, as the Palivizumab® (Synagis, MedImmune) [4].
The Palivizumab® is a humanized murine monoclonal antibody produced by recombinant technology that targets the A antigenic site of the F protein of HRSV. This drug demonstrates both neutralizing and fusion inhibitory activity and thus can prevent or reduce the severity of HRSV infection [5]. Palivizumab® has been used for some specific high-risk groups of children in Sao Paulo, Brazil, since 2007 [6].
Several laboratory techniques have been used in the diagnosis of HHRSV and among than Real-Time polymerase chain reaction (qPCR) can be used not only to detect the virus, but also to quantify them and try to do a parallel with the severity of HHRSV infection [7].
The aim of this study was to evaluate HHRSV detection through the currently available tests such as Direct Immunofluorescence Assay (DFA), Polimerase Chain Reaction (PCR) and Real-Time polymerase chain reaction (qPCR), among symptomatic children samples in use of Palivizumab®, and compare the viral load to another group of infected children without previous prophylaxis recruited from the community.
Study population and sample processing
Study group: The study group included infants with indication of palivizumab according to the health office normative resolution that was, children younger than 1 year of age, born with gestational age lower than 29 weeks, after hospital discharge, and children younger than 2 years, with hemodynamically unstable cardiopathy or chronic pulmonary disease, who needed treatment in the 6 months prior to the period of the HRSV season .Sixty nine nasopharyngeal aspirate samples from fifty eight children were collected from April to September 2008 every time they presented a new acute respiratory infection.
A questionnaire with the following epidemiological and clinical aspects for the study was applied: the time onset of respiratory symptoms and general symptoms, preexisting conditions, comorbidities, and risk factors such as contact with symptomatic people. Clinical aspects of this study have been already published elsewhere [8].
Control group: To analyze the viral load of HHRSV infected children in use of Palivizumab and HHRSV infected children not under prophylaxis with the drug, 18 nasopharyngeal aspirate samples from 18 outpatient children, previously diagnosed positive for HHRSV, were included in this study. This children population was described as part of another epidemiological study published previously.
This study was approved by the Ethics Committee of Sao Paulo Federal University (CEP 0342-08), and written consent was obtained from all patients or those responsible for the individual patient.
Samples were processed using previously published procedures [9].
Direct immunofluorescence assay
The DFA was performed using the kit “Simulfluor Respiratory Screen and Panel” (Chemicon Int., EUA), in accordance with the manufacturer’s instructions.
Molecular methods
The nucleic acid was extracted using the kit “QIAmp DNA Blood Kit” (Qiagen, Germany) following the manufacturer's recommendations.
For the PCR method, the primers used for detection of HHRSV target the conserved region of F gene [10].
For the qPCR amplification method, the reaction was done according to the protocol previously described by Homaira, et al. [11] targeting the gene P of HHRSV. The reactions were accomplished using the AgPath-IDTM One Step RT-PCR kit (Ambion, EUA), according to manufacturer instructions.
Statistical analysis
Statistical analyses were performed to evaluate the agreement among the different tests using Microsoft Office Excel software version 2007.
The t test was conducted to compare two means of continuous variables, with significance level of p < 0.05.
The mean age of children of the study group was nine months, ranging from three months to two years. Among them 92.8% (64/69) of samples were from children who were premature birth, 23.1% (16/69) had any kind of heart disease and 66.7% (46/69) any kind of pulmonary disease.
Eleven patients under prophylaxis had more than one sample collected. The time to onset of symptoms until sample collection date had an average and median of 3 days, ranging between one to seven days, the information was not available for one child in the study group. Four out of 11 patients had at least one positive sample, all patients with samples collected in different ARI episodes.
In this group 15.9% (11/69) of samples collected were positive for HHRSV in at least one technique. All samples were tested by the three techniques showing a positivity of 10.1% (7/69) for DFA, 13.0% (9/69) for PCR and a positivity of 14.5% (10/69) for qPCR. We observed overall concordances of 95.6% among DFA and qPCR, 94.2% among DFA and PCR and 95.6% among PCR and qPCR, with one sample exclusively positive by PCR and two samples exclusively positive by qPCR. Considering the detection according to the time of symptoms onset, molecular methods showed higher detection capacity in samples collected more than three days after symptoms onset (Figure 1).
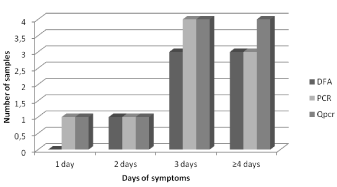
Figure 1. Distribution of the Respiratory Syncytial Virus detection by test according to the time of symptoms onset until the sample collection.
Samples that were negative for IF showed lower viral amount (average viral load = 4.0 log RNA copies/mL) than those with IF positive results (average viral load = 6.0 log RNA copies/mL) although no statistical difference between the averages (p = 0.17) was found. Similarly, PCR negative samples presented lower viral amount (average viral load = 3.8 log RNA copies/mL) when compared with those with positive results (average viral load = 5.7 log RNA copies/mL) (p = 0.072).
The four samples positive by qPCR and negative by DFA showed lower viral load values. These samples were collected from three children and they had received at least one Palivizumab® dose: one child with one dose, one child with two doses and another child with two episodes being one with two doses and the last episode with five doses.
Analysing HRSV positivity and number of doses received by children under survey, viral detection was more common among samples from children with fewer doses. The Figure 2 shows the number of positive samples obtained during HRSV survey and the cumulative doses received by children.
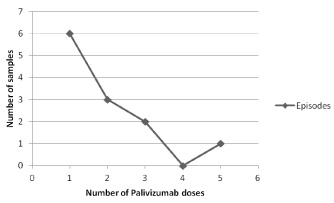
Figure 2. Cumulative number of doses of Palivizumab® received by children with positive samples for HRSV at the moment of collection.
Comparing the viral load of samples from children with or without use of Palivizumab®, there was no statistical difference; the average viral load was 5.1 log RNA copies/mL and 6.5 log RNA copies/mL respectively (Figure 3).
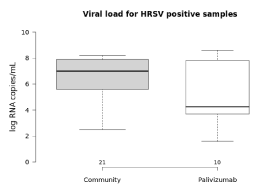
Figure 3. Comparative analysis of HRSV viral load between children in use of Palivizumab prophylaxis and children not under prophylaxis.
Among the eleven episodes of HRSV infection, seven were characterized as upper respiratory infection and four were bronchiolitis. One child was hospitalized due to bronchiolitis with total recovery after 10 days.
The comparison between the HRSV viral load on samples of children with and without bronchiolitis (p = 0.4) and according to the number of Palivizumab doses received showed no statistical significant differences.
Tables 1 and 2 showed the main characteristics of the study and control groups.
Table 1. Characteristics of Children under Palivizumab Prophylaxis.
Children under Palivizumab Prophylaxis |
Subject |
Time until symptoms onset (days) |
Age (years) |
Number of Palivizumab dosis |
Bronchiolitis |
Viral load (log copies/mL) |
01P |
2 |
0.75 |
1 |
Yes |
8.3 |
02P |
3 |
0.5 |
2 |
No |
8.6 |
05P |
6 |
0.41 |
1 |
No |
3.7 |
13P |
3 |
0.83 |
1 |
No |
7.8 |
15P |
4 |
0.66 |
1 |
Yes |
4.6 |
24P |
3 |
1 |
1 |
No |
4.4 |
25P |
1 |
1 |
2 |
No |
3.7 |
40P |
3 |
0.75 |
3 |
Yes |
4.1 |
61P |
4 |
1 |
3 |
Yes |
1.6 |
73P |
4 |
2 |
5 |
No |
3.9 |
Table 2. Characteristics of Children not in use of Palivizumab.
Children not under Palivizumab Prophylaxis |
Subject |
Time until symptoms onset (days) |
Age (years) |
Viral load (log copies/ mL) |
725 |
1 |
2 |
7.0 |
727 |
3 |
2 |
5.6 |
730 |
1 |
0.41 |
7.0 |
739 |
1 |
0.83 |
7.4 |
752 |
2 |
0.5 |
6.9 |
762 |
5 |
0.58 |
7.9 |
771 |
1 |
4 |
6.1 |
783 |
6 |
0.83 |
3.7 |
784 |
3 |
1 |
5.4 |
785 |
3 |
7 |
7.9 |
786 |
4 |
0.41 |
8.0 |
789 |
3 |
3 |
7.2 |
793 |
1 |
1 |
7.8 |
807 |
2 |
0.16 |
7.9 |
826 |
2 |
0.5 |
7.3 |
835 |
3 |
0.41 |
2.5 |
852 |
1 |
0.08 |
8.1 |
861 |
/ |
2021 Copyright OAT. All rights reserv
1 |
3.8 |
Analysing the parameters of age (average of 1.4 years) and time up to symptoms onset (average of 2.4 days), there were no statistical significant difference comparing the study and control group.
The aim of this study was to evaluate HRSV detection in high-risk children in use of Palivizumab® and verify the possible differences in viral load between groups of children with and without prophylaxis with Palivizumab.
Several studies showed that the Palivizumab® does not just prevent HRSV infection but also decreases the hospitalization occurrence [12], and in fact, only one of our study group was hospitalized. Despite the recognized importance of Palivizumab® prophylaxis, in our study 17.4% of children were infected by HRSV.
The observation of different techniques results showed that molecular methods had a superior detection rate when compared with immunofluorescence, mainly in samples of patients with three days of symptoms onset or more, as expected [13]. Indeed, the performance of qPCR was even better than the capacity of detection by PCR, particularly in cases of decreased viral shedding, as demonstrated with the comparison of viral loads, lower in those samples that could not have HRSV detected by DFA or by PCR.
The surveillance of children receiving prophylaxis with Palivizumab®, by quantifying the viral load obtained similar results when compared to those infected children without prophylaxis. In addition, the four cases with severe disease were also comparable in terms of viral load.
These data highlight the need to better understand the role of monoclonal antibody on the disease dynamic and viral replication since the viral load was not a good predictor of severity during the infection [14].
The authors declare that there is no conflict of interest. Despite the partial supporting of Sanofi Aventis Pasteur the authors state that the results of the present study are independent of any company interest.
- Gardner PS, Turk DC, Aherne WA, Bird T, Holdaway MD, et al. (1967) Deaths associated with respiratory tract infection in childhood. Br Med J 4: 316-320. [Crossref]
- Kesson AM (2007) Respiratory virus infections. Paediatr Respir Rev 8: 240-248. [Crossref]
- Szabo SM, Gooch KL, Bibby MM, Vo PG, Mitchell I, et al. (2013) The risk of mortality among young children hospitalized for severe respiratory syncytial virus infection. Paediatr Respir Rev 13 Suppl 2: S1- S8.
- No authors (1998) Palivizumab®, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-HRSV Study Group. Pediatrics 102: 531-537.
- Sánchez PJ (2002) Immunoprophylaxis for respiratory syncytial virus. Pediatr Infect Dis J 21: 473-478. [Crossref]
- SUS-SP (2007) Norma técnica relativa às diretrizes para a prevenção da infecção pelo virus sincicial respiratório.
- Fuller JA, Njenga MK, Bigogo G, Aura B, Ope MO, et al. (2013) Association of the CT values of real-time PCR of viral upper respiratory tract infection with clinical severity, Kenya. J Med Virol 85: 924-932.
- Monteiro AI, Bellei NC, Sousa AR, dos Santos AM, Weckx LY (2014) Respiratory infections in children up to two years of age on prophylaxis with palivizumab. Rev Paul Pediatr 32: 152-158. [Crossref]
- Bellei N, Carraro E, Perosa A, Granato C (2007) Patterns of influenza infections among different risk groups in Brazil. Braz J Infect Dis 11: 399-402. [Crossref]
- Erdman DD, Weinberg GA, Edwards KM, Walker FJ, Anderson BC, et al. (2003) GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J Clin Microbiol 41: 4298-4303. [Crossref]
- Homaira N, Luby SP, Petri WA, Vainionpaa R, Rahman M, et al. (2012) Incidence of respiratory virus-associated pneumonia in urban poor young children of Dhaka, Bangladesh, 2009-2011. PLoS One 7: e32056.
- Stewart DL, Ryan KJ, Seare JG, Pinsky B, Becker L, et al. (2013) Association of HRSV-related hospitalization and non-compliance with Palivizumab® among commercially insured infants: a retrospective claims analysis. BMC Infect Dis 13: 334.
- Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M (2009) Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl Infect Dis 11: 298-303.
- Landry ML, Ferguson D (2000) SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol 38: 708-711.



