Several theories for the origin of life have gained widespread acceptance, led by primordial soup, chemical evolution, metabolism first, and the RNA world. However, while new and existing theories often address a key step, there is less focus on a comprehensive abiogenic continuum leading to the last universal common ancestor. Herein, I present the “minimotif synthesis” hypothesis unifying select origin of life theories with new and revised steps. The hypothesis is based on first principles, on the concept of selection over long time scales, and on a stepwise progression toward complexity. The major steps are the thermodynamically-driven origination of extant molecular specificity emerging from primordial soup leading to the rise of peptide catalysts, and a cyclic feed-forward catalytic diversification of compounds and peptides in the primordial soup. This is followed by degenerate, semi-partially conservative peptide replication to pass on catalytic knowledge to progeny protocells. At some point during this progression, the emergence of RNA and selection could drive the separation of catalytic and genetic functions, allowing peptides and proteins to permeate the catalytic space, and RNA to encode higher fidelity information transfer. Translation may have emerged from RNA template driven organization and successive ligation of activated amino acids as a predecessor to translation.
abiogenesis, chemical evolution, peptide, first principles, minimotif, natural selection
Abiogenesis and the chemical evolution theory of life are generally accepted by scientists [1]. A central tenet of life is the coordination of proteins in cells to perform essential processes such as cell division and metabolism. Cellular processes are built from evolutionarily tuned sets of long-lived complexes (binding events) and shorter-lived complexes that produce chemical reactions (reaction events). These chemistries have exquisite specificity that may have evolved from the most ancestral life forms. At the most fundamental level, all origin-of-life hypotheses rely on exquisitely specific molecular recognition. If the specificity for molecular recognition were not manifested in primordial events relating to chemical evolution, I contend that it is highly unlikely that life could have arisen from the vast sea of weaker affinity competing, non-specific molecular interactions.
Until life can be recreated de novo, all origin of life theories can be considered inconclusive. However, all provide insight into pathways that may have led to the first replicating cells. Herein, I present several new ideas that expand upon origin of life theories and present alternative hypotheses as a basis for further exploration. These ideas are largely based on my laboratory's work on minimotifs, short contiguous peptide sequences with a known molecular functions. These ideas are discussed in the context of, and are consistent with many of the other well-established origin-of-life theories.
Modern complex protein folds and efficient enzymes probably did not just appear from the primordial soup. While some protein-protein interactions bury surfaces of >1000 Å2, these interactions likely arose through a stepwise process of mutation and selection over time. These favorable interactions may have provided a competitive advantage to protocells, self-organizing membrane bound structures with some properties of life. I ask: How did molecular recognition originate, become inherited, and evolve?
Herein, I explain the new Minimotif Synthesis hypothesis expanding upon existing theory and explaining how the primordial soup may have led to a ribonucleic acid (RNA) world [2-4]. The hypothesis includes new ideas about the evolution of molecular recognition and the abiogenic evolution of heredity peptides as predecessors of the RNA world (Figure 1). The minimotif synthesis is based on first principles, on the concept of selection over long time scales, and on a stepwise progression toward complexity. Additionally, it connects several gaps between other models. It can be considered an expansion, a differing perspective, and a consolidation of other models [3,5-9].
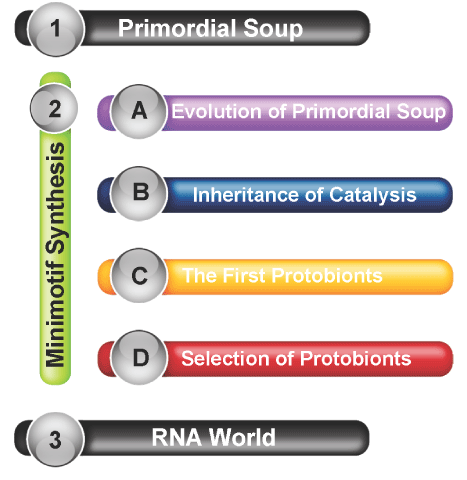
Figure 1. Minimotif synthesis in origin of life models. A diagram of how the general parts of the minimotif synthesis hypothesis explain the gap between the primordial soup and RNA world hypotheses.
Evolution of the primordial soup
Prebiotic Earth had several potential sources of extant biological molecules. The primordial soup experiments by Urey and Miller showed that, under reducing and neutral environmental conditions with carbon dioxide (CO2) and nitrogen (N2) likely existing on primitive Earth, many of the chemical constituents of life are formed leading to the abiogenesis hypothesis [10-13]. Other suspected sources of carbon molecules include carbonaceous chondrite meteorites and surface-based synthesis in hydrothermal vents [14-16]. I propose several modifications of the chemical evolution theory as detailed below [8]:
Thermodynamic- and selection-based origins of molecular interaction specificity: How did the extant sophisticated molecular recognition in biological systems originate?
I propose that the first life forms must have captured existing specific molecular interactions from the primordial soup. The originating specificity in life forms may have arisen from the inherent physiochemical properties of molecules to prefer interaction with a limited set of other molecules, as previously suggested for nucleic acids [17] (Figure 1, part I). Proteins also have a wide breadth of chemical moieties for molecular recognition. Indeed, as an example, it is easy to recognize that a carboxylic acid side chains of aspartic acid (Asp) or glutamic acid (Glu) have relatively high preference for interacting with a guanidinium moiety of arginine (Arg), which provides both stabilizing ionic and hydrogen bonding interactions [18].
Chemical evolution is proposed to have transformed the primordial soup [19-21]. I propose that natural selection of strong affinity intermolecular interactions was a driving principle in chemical evolution. Molecules with strong enthalpy-driven affinity for binding partners would restrict molecular motion, thus decreasing then entropy of each molecule, and thereby increasing stabilization. Evolution of primordial soup over long timescales could yield molecular partners stabilized in such a manner, which could have persisted and become more concentrated than those molecules lacking strong stabilizing interactions. Thus, thermodynamic forces over a long timescale may have drove the selection of molecules with strong affinities for other molecules. This selection may also have involved selection of chiral forms from racemic mixtures of optical isomers, potentially seeded stochastically [22,23]. Chiral seeding may have extraterrestrial origins, since some meteorites have an enantiomeric excess of L-amino acids. Note that this hypothesis is based upon the well-accepted first principles and selection.
Selection for strong affinity peptides: As part of the evolution of primordial soup, reasonable evidence supports the abiotic polymerization of amino acids into short peptides [9,24-30]. For example, short peptides may have formed by the salt-induced peptide formation mechanism [31]. Several mechanisms could have generated other bio-oligomers such as RNA [6,26,27,32]. Abiotic synthesis of biopolymers is challenging. However, the caveats of biopolymer formation in origin of life theory are extensively addressed in the literature and for the purposes of minimotifs synthesis hypothesis I assume the presence of abiotic biopolymers.
I focus on peptides for several reasons: (1) the evidence that the soup may have contained amino acids and peptides; (2) the strong affinities shown by extant peptides and proteins for many different types of small molecules; (3) the plethora of different types of catalytic activities in proteins; (4) the lack of strong secondary structures in peptides; and (5) the higher chemical stability of peptide bonds over RNA phosphodiester bonds, which are prone to hydrolysis. Although I focus on peptides, the evolution of RNA or other polymers may have followed a similar abiotic path and the concurrent prebiotic co-evolution of RNAs and proteins cannot be ruled out.
Many of the stable intermolecular interactions in primordial soup (Figure 1, Part I) were probably captured as binding interactions of amino acid R-groups in short peptides with other molecules present in the primordial soup (Figure 1, Part II). Likewise, I expect that physiochemical and thermodynamic forces also drove the affinity of peptides for other molecules. Unlike the simple chemical interaction examples given in Figure 1, Part I, a short peptide of just 2-3 amino acids can encode strong-affinity and high-specificity molecular interactions with small compounds. There are 100,000s of such interactions in the Minimotif Miner database and many of these peptides are modular and function outside of the context of the full-length protein e.g. [33-38]. In my lab, short, contiguous peptide elements in proteins with a known function, such as binding a small molecule or other protein, are called minimotifs [39,40]. Also known as short linear motifs (SLiMs), these minimotifs are extensively involved in molecular recognition [40]. In support of my hypothesis, minimotifs are also targets of evolution with subsets under negative and positive selection [41]. Minimotifs are also reproducibly discovered by simulated evolution [42].
If we assume that a particular primordial soup contained 10 natural amino acids [43] and, conservatively, only short peptides of less than 5 amino acids each, then there are ∼110,000 possible peptide sequence combinations. This pool may have been more expansive as abiotic synthesis of longer peptides is feasible and reanalysis of Urey and Miller’s original samples identified 23 amino acids [44]. This or a similar heterogeneous pool of peptides would provide a source for selection for specific binding to other molecules (small chemicals, mineral surfaces). For example, many short peptides can bind other molecules outside of the context of the complete folded protein [33-35,37].
Furthermore, although demonstrated for a longer peptide, screening a random peptide library identified 20 peptides that bind streptavidin with dissociation constants (Kds) as low as 5 nM [45,46]. Peptides that could tightly bind to a target molecule, and/or form more stable secondary structures would have generally been more constrained, and thus more thermodynamically stable. Over long timescales, these peptides would survive longer than those with weak affinities for targets or no stable secondary structure. Thus, peptides with strong affinity ligand interactions would be favored at the expense of those that don't form stable complexes. This proposed modification of the chemical evolution model provides an abiotic selection mechanism for specific sets of peptides that bind to other compounds such as sugars, amino acids, metabolites, nucleic acids, etc. The same selection/stabilization principle would also apply to peptides that bind to each other as well. Again, this portion of the model is based on first principles and the process of selection.
Catalytic functionalization of peptides: The emergence of catalysis is a premise of the metabolism-first model [47]. Once peptides evolved to recognize molecules, some could possess weak catalytic activities that break or form covalent bonds, thus becoming primitive catalysis (Figure 1, part III). In fact, many short peptides are known to possess catalytic activities [see reviews [48–50]]. For example, the Pro-x-x-Phe tetrapeptide (Pro=proline, Phe=phenylalanine, x=any amino acid) possesses catalytic aldol condensation activity for acetone with p-nitrobenzaldehyde [51]. Even shorter peptides, such as Pro-Pro, have aldol condensation activity [52] and many other chemistries are catalyzed by short peptides [48-50].
Reaction products would be chemically distinct from the substrates, thus would likely have a less optimal interaction with the peptide catalyst. Since these catalysts were peptides and lack the intricate binding pocket of many enzymes, they many have acted on classes of compounds, rather than on specific molecules [53,54]. These primitive catalysts may represent an early predecessor step of more complex and efficient enzymes [55]. There is also evidence supporting the hypothesis that short RNAs may also have emerged as weak catalysts [56].
Boundless propagating abiotic diversification of catalysts and molecules: As part of chemical evolution, I propose catalyst evolution through chemical synthesis. Catalytic peptides could have drastically altered the chemical composition of the primordial soup, concentrating certain chemicals, depleting others, and increasing the overall molecular complexity. The evolution of primitive catalysts could provide a mechanism for generating new molecules including new types of amino acids, peptides, and possibly nucleic acids. The genesis of new peptides could give rise to new types of catalysts, akin to a concept called autocatalytic sets [7]. A feed-forward cyclic process could ensue wherein peptides catalyze the formation of newer catalysts that spawn new molecules and new peptide catalysts, gradually increasing the molecular and catalytic complexity of the primordial soup (Figure 1, part IV). This scenario could set up an entropy-driven propagating abiotic diversification and expansion of more complex catalysts and more types of molecules, eventually leading to primitive metabolic pathways. Many of the molecules in the glycolytic pathway may have arisen abiotically [57]. This expansion may be boundless, initially abiotic and still ongoing in living organisms, with new expansions of enzymes into the new catalytic spaces.
At this point in the proposed model, the primordial soup would contain sets of peptides with strong affinity for small molecules and other sets of catalytic peptides. Two peptides, one with a binding activity and another with a catalytic activity, could fuse by peptide condensation reactions. Small peptides, such as Ser-His (serine-histidine) and Gly-Gly (Gly = glycine), can catalyze peptide bond formation [58,59]. This process could further drive the catalysts' generation cycle, ultimately producing primitive protein folds and enzymes that possessed both binding and catalytic activities (Figure 1, part III). Lupas et al. suggests this type of model, pointing out that it is supported by the observation that different extant proteins contains similar sequences and structural repeats [60].
At some point in the above chemical and catalyst evolution, possibly at the earliest stages, peptides, perhaps working cooperatively catalyzed the synthesis of nucleic acids and other polymers such RNA. Peptides cooperating in catalysis have been observed and would provide a much broader depth of catalytic capability [55]. Like peptides, RNA served both a dual binding and catalytic function. However, I favor the emergence of RNA at later stages, given the apparent lack or breadth of catalytic diversity in extant RNAs where extant RNA catalysis is focused primarily on nucleic acids substrates and that abiotic synthesis of nucleic acids is difficult. Peptides and proteins have a much broader repertoire of catalytic reaction types.
This expanded view of chemical evolution need not be compartmentalized; however, for concentration of compounds and activities, encapsulation into self-organizing protocells is feasible. I suggest that some or many of the aforementioned steps could also have occurred within a bounded protocell type system as well, which is addressed later.
A central concept applied so far in origin of life research is based on the premise that if synthesis of a compound under prebiotic conditions occurred, then it is feasible to have played a role in prebiotic evolution. Considering that the time scale of the above events may be more than a billion years, any system that propagates molecular and catalytic diversity, as I have proposed, could explain abiotic synthesis of many of the molecules of life. I offer that a catalytic propagation model would be favored.
As noted by Orgel, the puzzle is how we get from a soup of prebiotic organic molecules to the RNA world [61]? Others have questioned this as well, suggesting that genetic systems simpler than RNA may have served as a predecessor [8,24,62-65]. In contrast to the sophisticated high-fidelity nucleic acid-based inheritance observed in extant organisms and proposed in the RNA world, I hypothesize a lower fidelity predecessor where a simpler, less-exact stepwise process gave rise to the first hereditary information system.
In early protocells, or life forms it was important to pass on the catalytic information needed for metabolism. At the onset of life, it would seem beneficial to have catalytic and hereditary functions embodied in the same molecule such that specific molecular recognition and catalytic capability could be inherited together. Since ribozymes are catalytic, RNA could have served a dual function - as both a catalyst for reactions and as the hereditary material [66]. This is an attractive feature of the RNA world model. In support of this model, Cech noted: “it is unclear how a self-replicating system based on completely unrelated chemistry could have been supplanted by RNA” [65]. I consider the inverse: How could a catalytic system based on RNA be almost completely supplanted by proteins? Is it not possible that peptides were the original catalysts and, like RNA, had a second hereditary function that was supplanted by RNA? Many have suggested a predecessor of the RNA world, so herein, I focus on evidence consistent with the minimotif synthesis hypothesis. However, it is entirely possible that peptides as catalysts, and RNA as hereditary material, were born of abiotic chemical evolution and then converged into a translational system later in protocells and biopoesis of the first life forms.
As long as a template could be used to self-replicate peptide catalysts, nothing more is required for the genesis of peptide-based inheritance of molecular recognition and catalysis. Proteins clearly have a much wider breadth and efficiency as catalysts than RNAs. There is evidence that peptides could have had such templates, in that many extant protein structures and protein-protein interactions show secondary structure interactions with degeneracy. Those peptides that formed peptide-peptide interactions may have served as the initial pre-genomic genetic material. This hypothesis is akin to the Graded Autocatalysis Replication Domain (GARD) model previously proposed for chemical inheritance, but has the advantage of a potential replication mechanism [67].
I designate these peptide replicons as “minimotifosomes.” A minimotifosome is composed of two complementary peptides that bind each other and could, when separated, give rise to two peptides - each capable of serving as a template for the other in chemical synthesis (akin to base-pairing in nucleic acids). The amino acids aligned on the template through side chain interactions could be covalently joined forming peptide bonds with the aid of a catalyst, similar to the mechanism of semiconservative DNA replication (Figure 2A). In fact, self-replicating peptides and actions of peptides as templates have already been experimentally demonstrated for an α-helix [68-71]. Alternatively, as previously hypothesized, peptides could have replicated through a autocatalytic cyclic network involving two our more peptides [72]. Furthermore, several small peptides, such as Ser-His and Gly-Gly, have been shown to catalyze peptide bond formation [58,59]. These peptides could have served as a primitive form of the non-ribosomal peptide synthesis prevalent in modern life [see Norine tabase [73].
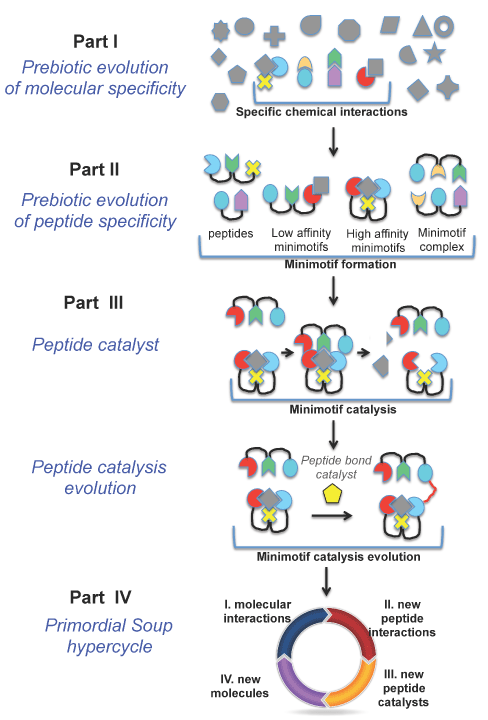
Figure 2. Minimotif synthesis model for peptide catalyst evolution from primordial soup. In Part I, stable interactions occur between compounds and amino acids, and among amino acids. The grayed figures are compounds and the colored compounds are amino acids. In Part II, peptide:compound and peptide:peptide complexes are more stable than uncomplexed peptides. Te most stable complexes, and molecules emerge out of the primordial soup. In Part III, some peptides or peptide complexes produce the first catalysts. These catalysts evolved, producing new molecules leading to new molecular interactions and new peptide interactions, and thus new catalysts in a cycle (parts III and IV).
Inger et al. previously suggested the GARD model for inheritance of chemical information in support of Oparin's metabolism first hypothesis [47,67,74]. However, one limitation noted was that such a system is incapable of evolution [75]. My model of minimotifosome replication provides an intuitive solution to this problem. These minimotifosomes could have had dual functionality, serving both as primitive catalysts and as the hereditary material, thus providing a simpler, primitive means for inheritance of catalytic capabilities.
The semi-conservative replication of minimotifosomes need not be perfect to provide a mechanism to pass on chemical catalysis information and to allow for mutation, selection, and thus, evolution. This process could be called “degenerate semi-conservative replication”, because minimotifosome replication may lack high fidelity. The known self-replicating peptides are α-helical, thus would be able to preserve intermolecular minimotif-minimotif contact residues such as the leucine (Leu) residues conserved in a leucine zipper (Figure 2A), but would have generally lower fidelity for the non-contact amino acids in the two helices [71]. There are other secondary structure interactions, such as β strand-β strand interactions [76,77], that could also produce low-fidelity replication. The lack of fidelity for non-contact amino acids could have been the first genetic source of mutation and genetic diversity, and acted upon by selection.
Some random coil minimotifs might have offered the highest genetic fidelity. For example, it is easy to envision how an Arg-Glu-Arg-Arg-Glu/Glu-Arg-Glu-Glu-Arg minimotifosome bound by salt bridges would encode molecular specificity; this implementation of complementary peptides serving as genetic material was suggested as a possibility by Orgel [61]. A subset of such random coils could have been primitive genetic information with mutation occurring by degenerate recognition. Inheritance does not likely need to be nearly as perfect as that current encoded by nucleic acids. However, the peptides do need to maintain enough fidelity to pass on the relevant information. In the case of nucleic acids, there is a critical error threshold at about 50% sequence identity to stably pass on hereditary information [19,64]. Notably, this would be different for peptides, likely requiring that only the key catalytic and binding residues be inefficiently passed on, also allowing for variation and evolution.
Encapsulation of chemicals, minimotifosomes, and minimotifs into fatty acid bound vesicles may have started at any earlier stage in the aforementioned progression. I do not propose any change to the potential role of protocells in the origins of life; rather, I point out how the minimotif synthesis hypothesis is consistent with protocell formation. Protocells form when self-organized lipid- or fatty-acid membranes encapsulate bio-oligomers and small molecules which, in the metabolism first model, are thought to be the predecessors to cells [47,74,78,79]. Protocells containing sets of peptides, minimotifosomes, and chemicals may have been the first prebiotic entities capable of metabolism, growth, and reproduction.
Lipids and fatty acids are capable of self-organizing into vesicles, and prebiotic synthesis of fatty acids is suspected [80,81]. Spontaneously formed bilayer vesicles could capture random collections of peptides, molecules, and minimotifosomes [78,82,83]. Semi-permeable membranes would allow small molecules to enter and exit the protocells through diffusion [81,84]. Yet the larger peptides, and minimotifosomes would be sequestered inside the protocells, thus encapsulating the genetic and catalytic material. Protocells are capable of division, spontaneously driven by the incorporation of fatty acids or lipids into existing bilayers [79,85,86]. Thus, catalytic peptides involved in fatty acid synthesis could have driven protocells division. Protocells with a selective advantage, such as those with better-performing catalysts, would dominate an ecosystem as previously shown in a experimental model system [58,87]. Thus, those protocells with favorable catalytic competencies encoded in minimotifosomes would outcompete other protocells for available resources.
While the minimotif synthesis hypothesis suggests minimotifosomes as the first carriers of hereditary information, there are severe limitations of minimotifosomes that would have led the use of nucleic acids as hereditary material: (1) the minimotifosome's length was probably constrained to short peptides because tertiary folds would interfere with replication; (2) if some minimotifs were proteolytic, this could minimotifosomes compromising the genetic material; (3) peptide duplex recognition would have been degenerate; and (4) only those minimotif duplexes that could serve as a template for self-replication could have served as primitive minimotifosomes.
Proteins are more efficient and have a much wider catalytic diversity when compared to peptides To fully take advantage of the catalytic protein space with longer and more complex proteins, and more efficiently preserve chemical catalysis information, eventually, the catalysis function would need to be separated from hereditary function. If the functions were separated this would allow protein folds to dominate, as peptide lacking tertiary structure would not longer be a restriction for replication. As proteins have more catalytic prowess, this would be a selective advantage. From the perspective of separating the hereditary information to another molecule, nucleic acids are obviously superior at encoding genetic information without degeneracy, thus chemical information could be passed on more efficiently. This likely drove the need for a different molecule as the genetic material (e.g. RNA).
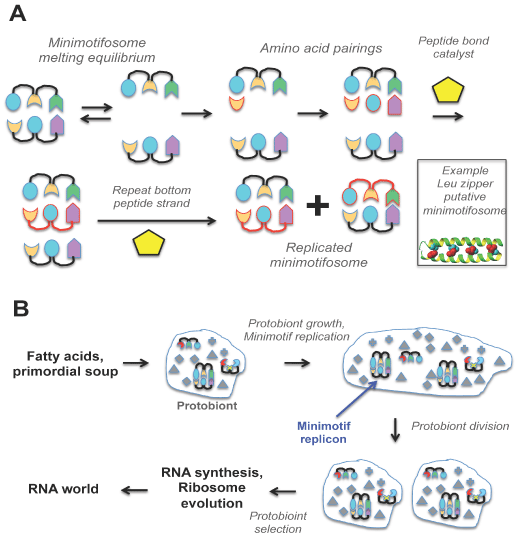
Figure 3. Degenerate semi-conservative minimotifosome replication model and its role in prebiotic evolution. A. Minimotifosome replication. The colored figures are amino acids. The figures with a red outline are newly synthesized peptides and are connected by red lines representing new peptide bonds. A structure of a leucine zipper is an example of an extant minimotif representing what a minimotifosome may have looked like. B. Lipids and/or fatty acids form protocells encapsulating peptides, minimotifosomes, and small molecules. Those protocells with sets of minimotifs capable of template-driven reproduction provide a basis for selection and evolution. Those protocells capable of fatty acid synthesis catalyze growth of the protocell leading to spontaneous division. Division of heterogeneous protocells leads to selection and, ultimately, to the birth of the ribosome and the RNA world.
The use RNA as the genetic material to encode peptides presents a major bottleneck in translating the genetic information of RNA into the catalytic information of proteins. A chicken and egg paradox noted for the RNA world hypothesis is that a RNA replicase function is needed prior to RNA becoming the genetic material [64]. Another similar paradox is that an RNA that replicates is proposed to have template driven replication within a double stranded segment of RNA. A helicase or high temperature would be need to stably melt the duplex allowing accessibility to the template for replication. Thus, helicase and the equivalent of single strand binding proteins would be needed to stabilize the template for primitive transcription and translation. However, these functions could not be encoded by proteins until after the origins of translation.
I propose a highly speculative stepwise mechanism that is largely adapted from the coding coenzyme handles hypothesis for evolution of the genetic code [88]. As discussed previously, early protocells may have contained both minimotifosomes and RNA. The RNA was likely in the form of more stable short hairpins and short double stranded duplexes, with some of each being ribozymes. The short RNAs having base pairing were likely more thermodynamically stable, due to constraints on molecular motion as discussed for short peptides above.
Short RNAs, possibly hairpins were likely selected as early genetic material, as longer RNAs likely had strong secondary structures that prevented their use as templates. Some of these short single stranded RNAs first served as templates to organize the predecessors of charged transfer RNAs (tRNAs) as previously suggested [88]. Upon ligation to form peptide bonds, the first RNA template-driven synthesis of peptides ensued. Some of these peptides were likely identical to the catalytic strand of minimotifosomes. Although minimotifosomes may have still served as the primary genetic material, the template driven synthesis and coenzyme handles hypothesis provides a route to early encoding of peptides by RNA. Protocells having this capability may have been able to more efficiently replicate the minimotifosome genetic material, provide a selective advantage to those possessing these activities.
However a mechanism to replicate the RNA would be need to give protocells a permanent advantage. Although there is no current evidence, once a minimotifosome or set of minimotifosomes working together acquired RNA replicase activity, this set the stage for inheritance of RNA encoding the peptides that previously were encoded by minimotifosomes. Through selection this would give a distinct advantage to those protocells. This would then a act through a feed forward mechanism, with the RNA template synthesis of peptides leading to the ribosome and the replication activity leading to more efficient RNA metabolism with polymerases, helicases, nucleic acid biosynthetic enzymes and other enzymes.
Although the order of the proposed steps can be questioned and the necessity for additional steps is likely relevant, the general notion of the interplay of minimotifosome coevolution with RNA
is attractive. Template-driven peptide synthesis may have led to the adoption of RNA as the hereditary material, and to evolution of a primitive ribosome. Ensuing events leading to the evolution of the ribosome and tRNAs have been previously addressed [88,89].
My minimotif synthesis hypothesis unifies some existing theory with new origin of life hypotheses that will need to be challenged, further developed, and experimentally tested.
This work was supported by the National Institutes of Health grants GM107983 and GM079689.
- Oparin AI (1976) Evolution of the concepts of the origin of life, 1924-1974. Orig Life 7: 3-8. [Crossref]
- Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418: 214-221. [Crossref]
- Kun Á, Szilágyi A, Könnyű B, Boza G, Zachar I, et al. (2015) The dynamics of the RNA world: insights and challenges. Ann N Y Acad Sci 1341: 75-95. [Crossref]
- Yarus M (2010) Life from an RNA world: the ancestor within Harvard Univ. Press, Cambridge, Mass.
- Bernal JD (1949) The physical basis of life. Proceedings of the Physical Society. Section A 62, 746–746.
- Cairns-Smith AG1 (2008) Chemistry and the missing era of evolution. Chemistry 14: 3830-3839. [Crossref]
- Kauffman SA (1993) The origins of order: self-organization and selection in evolution Oxford University Press, New York.
- Oparin A (1957) The origin of life on the earth. Academic Press Inc., New York.
- Rode BM1 (1999) Peptides and the origin of life. Peptides 20: 773-786. [Crossref]
- Cleaves HJ1, Chalmers JH, Lazcano A, Miller SL, Bada JL (2008) A reassessment of prebiotic organic synthesis in neutral planetary atmospheres. Orig Life Evol Biosph 38: 105-115. [Crossref]
- Miller SL, Urey HC (1959) Origin of Life. Science 130: 1622-1624. [Crossref]
- Miller SL, Urey HC (1959) Organic compound synthesis on the primitive earth. Science 130: 245-251. [Crossref]
- Miyakawa S, Yamanashi H, Kobayashi K, Cleaves HJ, Miller SL (2002) Prebiotic synthesis from CO atmospheres: implications for the origins of life. Proc Natl Acad Sci U S A 99: 14628-14631. [Crossref]
- Cronin JR, Pizzarello S (1983) Amino acids in meteorites. Adv Space Res 3: 5-18. [Crossref]
- Wächtershäuser G (1988) Before enzymes and templates: theory of surface metabolism. Microbiol Rev 52: 452-484. [Crossref]
- Pizzarello S, Shock E (2010) The organic composition of carbonaceous meteorites: the evolutionary story ahead of biochemistry. Cold Spring Harb Perspect Biol 2: a002105. [Crossref]
- Shapiro R (2006) Small molecule interactions were central to the origin of life. Q Rev Biol 81: 105-125. [Crossref]
- Mitchell JB1, Thornton JM, Singh J, Price SL (1992) Towards an understanding of the arginine-aspartate interaction. J Mol Biol 226: 251-262. [Crossref]
- Eigen M (1971) Molecular self-organization and the early stages of evolution. Experientia 27: 149–212.
- Spiegelman S (1971) An approach to the experimental analysis of precellular evolution. Q Rev Biophys 4: 213-253. [Crossref]
- Follmann H, Brownson C (2009) Darwin's warm little pond revisited: from molecules to the origin of life. Naturwissenschaften 96: 1265-1292. [Crossref]
- Clark S (1999) Polarized starlight and the handedness of life. American Scientist 87: 336.
- Córdova A, Engqvist M, Ibrahem I, Casas J, Sundén H (2005) Plausible origins of homochirality in the amino acid catalyzed neogenesis of carbohydrates. Chem Commun (Camb) : 2047-2049. [Crossref]
- Cairns-Smith AG (1982) Genetic takeover and the mineral origins of life Cambridge University Press, Cambridge?; New York.
- Carny O, Gazit E (2005) A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J 19: 1051-1055. [Crossref]
- Ferris JP1, Hill AR Jr, Liu R, Orgel LE (1996) Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381: 59-61. [Crossref]
- Fox SW (1995) Thermal synthesis of amino acids and the origin of life. Geochim Cosmochim Acta 59: 1213-1214. [Crossref]
- Fox SW, Harada K, Kendrick J (1959) Production of Spherules from Synthetic Proteinoid and Hot Water. Science 129: 1221-1223. [Crossref]
- Fox SW, Harada K, Vegotsky A (1959) Thermal polymerization of amino acids and a theory of biochemical origins. Experientia 15: 81-84. [Crossref]
- Shapiro R (1987) Origins: a skeptic’s guide to the creation of life on earth Bantam Books, Toronto; New York, USA.
- Rode BM & Suwannachot Y (1999) The possible role of Cu(II) for the origin of life. Coordination Chemistry Reviews 190-192: 1085–1099.
- Danger G, Plasson R, Pascal R (2012) Pathways for the formation and evolution of peptides in prebiotic environments. Chem Soc Rev 41: 5416-5429. [Crossref]
- Thiele A, Zerweck J, Weiwad M, Fischer G, Schutkowski M (2009) High-density peptide microarrays for reliable identification of phosphorylation sites and upstream kinases. Methods Mol Biol 570: 203-219. [Crossref]
- Wang Z (2009) The peptide microarray-based assay for kinase functionality and inhibition study. Methods Mol Biol 570: 329-337. [Crossref]
- Wu C, Li SS (2009) CelluSpots: a reproducible means of making peptide arrays for the determination of SH2 domain binding specificity. Methods Mol Biol 570: 197-202. [Crossref]
- Mi T, Merlin JC, De2021 Copyright OAT. All rights reserv (2012) Minimotif Miner 3.0: database expansion and significantly improved reduction of false-positive predictions from consensus sequences. Nucleic Acids Res 40: D252–260. [Crossref]
- Liu BA, Engelmann BW, Nash PD (2012) High-throughput analysis of peptide-binding modules. Proteomics 12: 1527-1546. [Crossref]
- Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, et al. (2009) Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res 37: D185-190. [Crossref]
- Balla S, Thapar V, Verma S, Luong T, Faghri T, et al. (2006) Minimotif Miner: a tool for investigating protein function. Nature Methods 3: 175–177. [Crossref]
- Vyas J, Nowling RJ, Maciejewski MW, Rajasekaran S, Gryk MR, et al. (2009) A proposed syntax for Minimotif Semantics, version 1. BMC Genomics 10: 360. [Crossref]
- Lyon KF, Strong CL, Schooler SG, Young RJ, Roy N, et al. (2015) Natural variability of minimotifs in 1092 people indicates that minimotifs are targets of evolution. Nucleic Acids Res 43: 6399-6412. [Crossref]
- Song T, Gu H (2014) Discriminative motif discovery via simulated evolution and random under-sampling. PLoS One 9: e87670. [Crossref]
- Miller SL (1987) Which organic compounds could have occurred on the prebiotic earth? Cold Spring Harb Symp Quant Biol 52: 17-27. [Crossref]
- Parker ET, Cleaves HJ, Dworkin JP, Glavin DP, Callahan M, et al. (2011) Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proceedings of the National Academy of Sciences 108, 5526–5531.
- Wilson DS, Keefe AD, Szostak JW (2001) The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci U S A 98: 3750-3755. [Crossref]
- Keefe AD, Szostak JW (2001) Functional proteins from a random-sequence library. Nature 410: 715-718. [Crossref]
- Oparin AI (1962) Origin and evolution of metabolism. Comp Biochem Physiol 4: 371-377. [Crossref]
- Brack A (2007) From interstellar amino acids to prebiotic catalytic peptides: a review. Chem Biodivers 4: 665-679. [Crossref]
- Tanaka F, Barbas CF 3rd (2002) A modular assembly strategy for improving the substrate specificity of small catalytic peptides. J Am Chem Soc 124: 3510-3511. [Crossref]
- Brack A1, Barbier B (1990) Chemical activity of simple basic peptides. Orig Life Evol Biosph 20: 139-144. [Crossref]
- Kofoed J, Nielsen J, Reymond JL (2003) Discovery of new peptide-based catalysts for the direct asymmetric aldol reaction. Bioorg Med Chem Lett 13: 2445-2447. [Crossref]
- 52 Krattiger P, Kovasy R, Revell JD, Ivan S & Wennemers H (2005) Increased structural complexity leads to higher activity: peptides as efficient and versatile catalysts for asymmetric aldol reactions. Org. Lett. 7, 1101–1103.
- Kacser H, Beeby R (1984) Evolution of catalytic proteins or on the origin of enzyme species by means of natural selection. J Mol Evol 20: 38-51. [Crossref]
- Szilágyi A, Kun A, Szathmáry E (2012) Early evolution of efficient enzymes and genome organization. Biol Direct 7: 38. [Crossref]
- Broo KS, Nilsson H, Nilsson J, Flodberg A, Baltzer L (1998) Cooperative Nucleophilic and General-Acid Catalysis by the HisH + -His Pair and Arginine Transition State Binding in Catalysis of Ester Hydrolysis Reactions by Designed Helix-Loop-Helix Motifs. Journal of the American Chemical Society 120: 4063–4068.
- Benner SA, Ellington AD, Tauer A (1989) Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci U S A 86: 7054-7058. [Crossref]
- Keller MA, Turchyn AV, Ralser M (2014) Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol Syst Biol 10: 725. [Crossref]
- Adamala K, Szostak JW (2013) Competition between model protocells driven by an encapsulated catalyst. Nat Chem 5: 495-501. [Crossref]
- Gorlero M, Wieczorek R, Adamala K, Giorgi A, Schininà ME, et al. (2009) Ser-His catalyses the formation of peptides and PNAs. FEBS Lett 583: 153-156. [Crossref]
- Lupas AN, Ponting CP, Russell RB (2001) On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol 134: 191-203. [Crossref]
- Orgel LE (1998) The origin of life--a review of facts and speculations. Trends Biochem Sci 23: 491-495. [Crossref]
- Joyce GF1 (1989) RNA evolution and the origins of life. Nature 338: 217-224. [Crossref]
- Orgel LE (2004) Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39: 99-123. [Crossref]
- Robertson MP, Joyce GF (2012) The origins of the RNA world. Cold Spring Harb Perspect Biol 4. [Crossref]
- Cech TR (2012) The RNA worlds in context. Cold Spring Harb Perspect Biol 4: a006742. [Crossref]
- Gilbert W (1986) Origin of life: The RNA world. Nature 319: 618–618.
- Inger A, Solomon A, Shenhav B, Olender T, Lancet D (2009) Mutations and lethality in simulated prebiotic networks. J Mol Evol 69: 568-578. [Crossref]
- Issac R, Ham YW, Chmielewski J (2001) The design of self-replicating helical peptides. Curr Opin Struct Biol 11: 458-463. [Crossref]
- Ghosh I, Chmielewski J (2004) Peptide self-assembly as a model of proteins in the pre-genomic world. Curr Opin Chem Biol 8: 640-644. [Crossref]
- Kauffman S (1996) Self-replication. Even peptides do it. Nature 382: 496-497. [Crossref]
- Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR (1996) A self-replicating peptide. Nature 382: 525-528. [Crossref]
- Lee DH, Severin K, Ghadiri MR (1997) Autocatalytic networks: the transition from molecular self-replication to molecular ecosystems. Curr Opin Chem Biol 1: 491-496. [Crossref]
- Caboche S, Pupin M, Leclère V, Fontaine A, Jacques P, et al. (2008) NORINE: a database of nonribosomal peptides. Nucleic Acids Res 36: D326-331. [Crossref]
- Oparin AI (1961) [The origin of life]. Nord Med 65: 693-697. [Crossref]
- Vasas V, Szathmáry E, Santos M (2010) Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc Natl Acad Sci USA 107: 1470–1475.
- Remaut H, Waksman G (2006) Protein-protein interaction through beta-strand addition. Trends Biochem Sci 31: 436-444. [Crossref]
- Maury CP1 (2015) Origin of life. Primordial genetics: Information transfer in a pre-RNA world based on self-replicating beta-sheet amyloid conformers. J Theor Biol 382: 292-297. [Crossref]
- Deamer DW, Barchfeld GL (1982) Encapsulation of macromolecules by lipid vesicles under simulated prebiotic conditions. J Mol Evol 18: 203-206. [Crossref]
- Zhu TF, Szostak JW (2009) Coupled growth and division of model protocell membranes. J Am Chem Soc 131: 5705-5713. [Crossref]
- Hargreaves WR, Mulvihill SJ, Deamer DW (1977) Synthesis of phospholipids and membranes in prebiotic conditions. Nature 266: 78-80. [Crossref]
- Monnard PA, Deamer DW (2002) Membrane self-assembly processes: steps toward the first cellular life. Anat Rec 268: 196-207. [Crossref]
- Bangham AD, Standish MM, Watkins JC (1965) Diffusion of univalent ions across the lamellae of swollen phospholipids. J Mol Biol 13: 238-252. [Crossref]
- Peretó J (2005) Controversies on the origin of life. Int Microbiol 8: 23-31. [Crossref]
- Chang TMS (2007) Artificial cells: biotechnology, nanomedicine, regenerative medicine, blood substitutes, bioencapsulation, cell/stem cell therapy World Scientific, Hackensack, NJ, USA.
- Stano P, Luisi PL (2010) Achievements and open questions in the self-reproduction of vesicles and synthetic minimal cells. Chem Commun (Camb) 46: 3639-3653. [Crossref]
- Budin I, Debnath A, Szostak JW (2012) Concentration-driven growth of model protocell membranes. J Am Chem Soc 134: 20812-20819. [Crossref]
- Chen IA, Roberts RW, Szostak JW (2004) The emergence of competition between model protocells. Science 305: 1474-1476. [Crossref]
- Szathmáry E (1993) Coding coenzyme handles: a hypothesis for the origin of the genetic code. Proc Natl Acad Sci U S A 90: 9916-9920. [Crossref]
- Fox GE (2010) Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2: a003483. [Crossref]



