The case of an identification of a small bone found in a blood sausage is described. The similitude with a rodent bone leads the consumer to demand the government organism responsible for food quality control. Bone and blood sausage DNA analysis was performed in order to determinate the origin species and the possibility of food contamination and/or adulteration. DNA sequence analysis confirmed the pig origin of both samples with an identity higher than 98%.
DNA sequencing, species identification, food contamination
Species identification of raw materials compounding a human consumption food may be informative in case of adulteration and/or contamination due to bad practices during the elaboration.
Mitochondrial DNA is a useful tool in forensic sciences because it helps to establish the source of a biological sample by means of linage identification [1]. The advantage of mitochondrial-based DNA analyses derives from the fact that there are several mitochondria per cell and many mitochondrial DNA molecules within each mitochondrion, increasing the possibility of obtaining results from critical samples [2]. The mitochondrial cytochrome b (cytb) gene is among the most extensively sequenced to date among vertebrates [3]. This gene has highly conserved nucleotide sequences which are known to be species specific and is widely analyzed for species specific identification, phylogenetic analysis and forensic investigations [4-6]. The method of species identification by direct sequencing using forensically informative nucleotide sequencing (FINS) of mitochondrial regions has been broadly used for identification of various kinds of animal species [7,8].
The blood sausage contains mainly pork blood and fat, onions, and salt. This sausage could be adulterated mixing meats and fats of diverse origins [9].
Here is described a case in which a bone, suspected to belong to a small animal like a rodent, was found inside a blood sausage (Figure 1). This finding leads the consumer to file a lawsuit against the government organism responsible for food quality control.
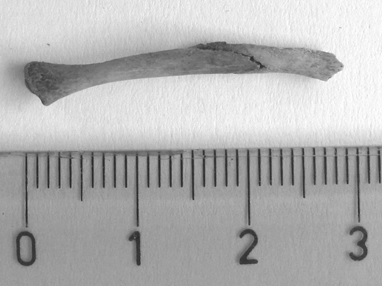
Figure 1. Piece of bone found inside the blood sausage used as evidence for the prosecutor (scale in cm).
DNA extraction
DNA was extracted from both the bone and the blood sausage. Bone was treated overnight with 0.5 M EDTA and 2% sarcosil. 0.5 M EDTA was changed every day during a week. After decalcification, DNA was obtained after a digestion with 0.25 mg/ml of proteinase K (50 mM Tris-HCL, 25 mM EDTA, 2% N-laurylsarcosine and 50 mM DTT) until complete tissue disaggregation, followed by an organic extraction with chloroform and ammonium acetate, and ethanol precipitation. Blood sausage DNA was obtained using Wizard® Genomics DNA Purification kit (Promega Corporation, WI, USA) according to the manufacturer´s instructions.
Cytb gene PCR reaction
Universal oligonucleot2021 Copyright OAT. All rights reserved to amplify by PCR a 358 bp fragment of the cytb gene [4]. The PCR was performed in 25 µl, using 10 ng of template DNA and 1,25 u of mi-Taq DNA polymerase (Metabion, Martinsried, Germany) according the manufacturer’s instructions. After 2 min of initial denaturation (94°C), 30 cycles were run each comprising 40 s denaturation (94°C), 40 s annealing (52°C), and 40 s extension (72°C), followed by a final extension of 5 min (72°C). Cow and human DNA samples were included as positive controls.
DNA Sequencing
Amplicons were purified by PEG precipitation (10% polietilenglycol 6000, 1.25% NaCl), centrifuged for 15 min a 15.000g, ethanol washed and resuspended in 15 µl of high purity water.
PCR products were sequenced using DYEnamic ET Dye Terminator Kit (GE Healthcare) and the universal cytb primers, using a MegaBACE 1000 automated sequencer (GE Healthcare). The DNA sequences were carefully reviewed using the Chromas 2.13 software (Helensvale, Queensland, Australia). The nucleotide sequences obtained were analyzed by applying blastn 2.2.24 against GenBank database.
There are several techniques of food components analysis. Usually, DNA analysis are preferred to other methods because are more specific, less expensive and less time consuming than e.g. mid-infrared spectroscopy, enzyme-linked immunosorbent assay or capillary electrophoresis [10]. Direct sequencing of PCR amplified fragment is, in the case of certain samples, the technique of choice since is accurate, simple and moderate cost. Our primary goal was to identify the species of origin of the bone found in the blood sausage. In addition, the species origin of components of the blood sausages was determined. For these reason, we amplified a fragment of a mitochondrial cytb gene, a highly conserved region among species.
A cytb gene fragment were successfully amplified from the samples and sequenced. All PCR products (358 bp) showed the expected size when separated on a 1.2% agarose gel. Therefore, the sequences were blasted to identify species against those with the highest similarities available in GenBank. Comparison of the bone DNA sequence reveal that the bone sample was from Sus scrofa (pig) with a 99% of identity. The blood sausage sample showed a 98% of identity with S. scrofa. The analysis of the blood sausage sequence with Chromas 2.32 software (Technelysium Pty Ltd., Tewantin, Australia) reveled that the sequence presented double peaks at certain positions probably due to a possible mix of ingredients (probably cow meat or blood).
This study reveals that the direct sequencing using FINS of mitochondrial regions was a useful tool for the species-level identification of the bone sample. Also it was possible to determine the main species present in the blood sausage. However, this methodology did not allow identify the origin of some components present in a low percentage in mixed samples as the blood sausage. In this case is recommended the use of specific primers in order to detect as few as 0.05% of one component in a mix [11]. The species-specific PCR suffers disadvantages with regard to a multiplicity of the species to be tested. It is impossible to design specific primer sets for all species that should be covered in forensic field [12].
- Pereira F, Carneiro J, van Asch B (2010) A Guide for Mitochondrial DNA Analysis in Non-Human Forensic Investigations. The Open Forensic Science Journal 3: 33-44.
- Walker JA, Hughes DA, Anders BA, Shewale J, Sinha SK, et al. (2003) Quantitative intra-short interspersed element PCR for species-specific DNA identification. Anal Biochem 316: 259-269. [Crossref]
- Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. J Mol Evol 32(2): 128-44. [Crossref]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, et al. (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers, Proc Natl Acad Sci USA 86: 6196-6200. [Crossref]
- Hsieh HM, Chiang HL, Tsai LC, Huang NE, Linacre A, Lee JC (2001) Cytochrome b gene for species identification of the conservation animals, Forensic Sci Int 122: 7-18. [Crossref]
- Linacre A, Lee JC (2005) Species determination: the role and use of the cytochrome b gene. Methods Mol Biol 297: 45-52. [Crossref
- Sahajpal V, Goyal SP (2010) Identification of a forensic case using microscopy and forensically informative nucleotide sequencing (FINS): A case study of small Indian civet (Viverricula indica). Sci Justice 50: 94-97.
- Gupta SK, Kumar A, Hussain SA, Vipin, Singh L (2013) Cytochrome b based genetic differentiation of Indian wild pig (Sus scrofa cristatus) and domestic pig (Sus scrofa domestica) and its use in wildlife forensics. Sci Justice 53: 220-2.
- Código Alimentario Argentino.
http://www.alimentosargentinos.gov.ar/programa_calidad/Marco_Regulatorio/CAA.asp
- Bottero MT, Dalmasso A (2010) Animal species identification in food products: Evolution of biomolecular methods. Vet J 190: 34-38. [Crossref]
- Ha JC, Jung WT, Nam YS, Moon TW (2005) PCR Identification of Ruminant Tissue in Raw and Heat-Treated Meat Meals. Journal of Food Protection 69: 2241-2247. [Crossref]
- Imaizumi K, Akutsu T, Miyasaka S, Yoshino M (2007) Development of species identification tests targeting the 16S ribosomal RNA coding region in mitochondrial DNA. Int J Legal Med 121: 184-191. [Crossref]

