Abstract
Retinaldehyde (retinal) has been identified as having superior topical activity, unrivaled topical bioavailability, and is less irritating than most other vitamin A derivatives. However, it has not received its deserved market attention possibly due to formulation-related issues. A novel compound, Retinaldehyde γ-Cyclodextrin Complex (RCC), has been developed to address the principal issues preventing retinaldehyde to enter the marketplace. Gene expression data of RCC have shown certain unique skin care attributes that are not present in retinaldehyde, including significantly less topical irritation.
Key words
retinaldehyde, retinoid, retinaldehyde cyclodextrin complex, cellular senescence, skin aging, rosacea, acne, topical wounds
Introduction
Retinaldehyde (Retinal) is gaining in popularity in anti-aging cosmetic products. It is well known that all forms of Vitamin A have positive effects for reducing fine lines and wrinkles of the face associated with skin aging [1]. The biology of skin aging process consists of two types; intrinsic or chronological aging, and photoaging. Retinoids have been popular for the treatment of skin aging and acne [2,3]. Amongst the retinoids, Tretinoin is possibly is the most widely investigated retinoid for photoaging and acne treatments [4]. However, irritant reactions such as burning, scaling and/or dermatitis associated with retinoid therapy limit their acceptance by patients. This problem seems more prominent with Tretinoin and Tazarotene whereas retinaldehyde and retinol seem to be less irritating. Novel drug delivery systems have been developed to minimize these side effects. In particular, nanoparticles have shown good potential in improving the stability, tolerability and efficacy of retinoids [5]. However, nanoparticle have developed certain less favorable consumer attention for their potential safety issues. Retinaldehyde has shown to be unstable in typical cosmetic formulations. This intrinsic instability has posed problems in the inclusion of retinaldehyde and other retinoids in topical anti-aging products [6-8]. The approaches to retinal stability include certain Schiff's bases with polylysine [9], retinaldehyde - hyaluronic acid fragments [10], and combinatorial treatments [11].
|
Gene ID
|
Gene Name
|
Retinol
|
Retinol
|
Retinaldehyde
|
RCC
|
Gene Function
|
|
0.5%
|
0.10%
|
0.10%
|
0.10%
|
|
DSC1
|
desmocollin
|
-11.89
|
-2.38
|
-6.97
|
-12.36
|
Extracellular matrix/Cell adhesion
|
|
TNF
|
tumor necrosis factor alpha
|
n/a
|
n/a
|
-3.78
|
-5.92
|
Inflammatory response, Extracellular matrix breakdown
|
|
FLG
|
epidermal filaggrin
|
-2.35
|
-0.47
|
-3.64
|
-5.60
|
Barrier function
|
|
LOR
|
loricrin
|
n/a
|
n/a
|
-3.82
|
-4.32
|
Keratinocyte differentiation/Barrier function
|
|
BMP4
|
bone morphogenetic protein 4
|
n/a
|
n/a
|
-2.16
|
-3.39
|
Whitening/Melanogenesis
|
|
DSG1
|
desmoglein 1
|
-5.33
|
-1.07
|
-2.26
|
-2.95
|
Extracellular matrix, Cell adhesion/Barrier function
|
|
CDSN
|
corneodesmosin
|
n/a
|
n/a
|
-2.57
|
-2.74
|
Barrier function
|
|
PTGS2
|
cyclooxygenase 2 (COX-2)
|
4.35
|
0.87
|
n/a
|
-2.60
|
Cytokine/Chemokine/Inflammatory response
|
|
LIF
|
leukemia inhibitor factor
|
10.93
|
2.19
|
n/a
|
-2.45
|
Inflammatory response
|
|
KRT1
|
keratin 1
|
-6.30
|
-1.26
|
n/a
|
-2.30
|
Keratinocyte differentiation
|
|
LCE3D
|
late cornified envelope 3D
|
n/a
|
n/a
|
-2.00
|
-2.15
|
Barrier function
|
|
NGF
|
nerve growth factor
|
-7.97
|
-1.59
|
n/a
|
-2.06
|
Inflammation/Collagen synthesis/Repair
|
|
IL6
|
interleukin 6
|
3.18
|
0.64
|
n/a
|
-2.04
|
Cytokine/Chemokine/Inflammatory Response
|
|
IL1A
|
interleukin 1 alpha
|
3.76
|
0.75
|
n/a
|
-2.02
|
Cytokine/Chemokine/Inflammatory response
|
|
IL8
|
interleukin 8
|
11.84
|
2.37
|
n/a
|
-1.92
|
Cytokine/Chemokine/Inflammatory response
|
|
MMP1
|
matrix metallopeptidase 1/collagenase
|
n/a
|
n/a
|
n/a
|
-1.82
|
Extracellular matrix breakdown
|
|
TGM1
|
transglutaminase
|
n/a
|
n/a
|
n/a
|
-1.70
|
Keratinization/Barrier function
|
|
BMP2
|
bone morphogenetic protein 2
|
4.44
|
0.89
|
n/a
|
-1.69
|
Whitening/Melanogenesis
|
|
HMOX1
|
hemeoxygenase 1
|
n/a
|
n/a
|
2.12
|
-1.49
|
Anti-oxidant/Oxidative Stress
|
|
VEGFA
|
vascular endothelial growth factor A
|
n/a
|
n/a
|
n/a
|
-1.47
|
Growth factor/Cell proliferation/Wound healing
|
|
MITF
|
micropthalmia-associated transcription factor
|
-1.85
|
-0.37
|
n/a
|
-1.46
|
Whitening/Melanogenesis
|
|
DSG3
|
desmoglein 3
|
1.99
|
0.40
|
1.92
|
1.68
|
Extracellular matrix, Cell adhesion/Barrier function
|
|
KLK8
|
kallikrein 8
|
n/a
|
n/a
|
2.13
|
1.88
|
Desquamation/Extracellular matrix breakdwon
|
|
ITGB4
|
b4 integrin
|
n/a
|
n/a
|
n/a
|
1.89
|
Cell adhesion/Barrier function
|
|
KLK5
|
kallikrein 5
|
n/a
|
n/a
|
n/a
|
1.96
|
Desquamation
|
|
TGFB1
|
transforming growth factor beta 1
|
2.04
|
0.41
|
2.20
|
2.17
|
Extracellular matrix intergrity, Cell adhesion/Barrier function
|
|
TP63
|
tumor protein p63; tp73-like
|
-1.41
|
-0.28
|
n/a
|
2.22
|
Cell cycle/proliferation
|
|
GSTT1
|
glutathione S transferase theta 1
|
n/a
|
n/a
|
n/a
|
2.44
|
Anti-oxidant/Oxidative Stress
|
|
KLK7
|
kallikrein 7
|
1.62
|
0.32
|
3.13
|
3.02
|
Desquamation/Extracellular matrix breakdwon
|
|
AQP3
|
aquaporin 3
|
n/a
|
n/a
|
4.85
|
5.96
|
Keratinocyte proliferation, Differentiation, Hydration
|
|
CLDN7
|
claudin 7
|
7.04
|
1.41
|
7.79
|
9.29
|
Barrier function
|
|
KLK6
|
kallikrein 6
|
13.49
|
2.70
|
11.28
|
12.99
|
Inflammation/Barrier function, Extracellular matrix breakdown
|
|
AQP10
|
aquaporin 10
|
-96.92
|
-19.38
|
n/a
|
n/a
|
Hydration
|
|
AQP7
|
aquaporin 7
|
-88.08
|
-17.62
|
n/a
|
n/a
|
Hydration
|
|
CAT
|
catalase
|
-2.34
|
-0.47
|
n/a
|
n/a
|
Anti-oxidant/Oxidative Stress
|
|
CLDN1
|
claudin 1
|
-1.74
|
-0.35
|
n/a
|
n/a
|
Barrier function
|
|
COL4A1
|
collagen IV a1
|
1.87
|
0.37
|
n/a
|
n/a
|
This gene encodes type IV alpha collagen chain
|
|
FGF2
|
|
1.92
|
0.38
|
n/a
|
n/a
|
Regulation of cell survival, cell division, angiogenesis, celldifferentiation and cell migration
|
|
HBEGF
|
heparin-binding EGF-like growth factor
|
7.73
|
1.55
|
1.90
|
n/a
|
Growth factor/Keratinocyte migration
|
|
IL1B
|
interleukin 1 beta
|
9.95
|
1.99
|
2.39
|
n/a
|
Cytokine/Chemokine/Inflammatory response
|
|
ITGB1
|
b1 integrin/Fibronectin 1 receptor
|
n/a
|
n/a
|
1.59
|
n/a
|
Cell adhesion/Barrier function
|
|
IVL
|
|
2.49
|
0.50
|
n/a
|
n/a
|
Component of keratinocyte crosslinked envelope
|
|
KRT10
|
|
-6.84
|
-1.37
|
n/a
|
n/a
|
This gene encodes a member of the type I cytokeratin family
|
|
KRT5
|
|
-3.03
|
-0.61
|
n/a
|
n/a
|
This type II cytokeratin is in the basal layer of the epidermis
|
|
MC1R
|
|
-2.12
|
-0.42
|
n/a
|
n/a
|
This gene encodes the receptor protein for melanocyte-stimulating hormone (MSH).
|
|
MKI67
|
|
-2.28
|
-0.46
|
n/a
|
n/a
|
This gene encodes a nuclear protein necessary for cellular proliferation
|
|
MMP3
|
|
2.85
|
0.57
|
n/a
|
n/a
|
This gene encodes an enzyme which degrades fibronectin, laminin, collagens III, IV, IX, and X, and cartilage proteoglycans
|
|
MT2A
|
|
2.76
|
0.55
|
n/a
|
n/a
|
MT2A (metallothionein 2A) is a protein-coding gene that bind various heavy metals
|
|
PCNA
|
|
-2.38
|
-0.48
|
n/a
|
n/a
|
Auxiliary protein of DNA polymerase delta involved in the control of eukaryotic DNA replication
|
|
PKP1
|
|
-2.04
|
-0.41
|
n/a
|
n/a
|
Plakophilin proteins involved in molecular recruitment and stabilization during desmosome formation
|
|
TNC
|
tenascin C
|
n/a
|
n/a
|
-1.73
|
n/a
|
Extracellular matrix
|
|
TP73
|
|
-2.85
|
-0.57
|
n/a
|
n/a
|
Participates in the apoptotic response to DNA damage.
|
|
TXN
|
thioredoxin
|
n/a
|
n/a
|
2.12
|
n/a
|
Anti-oxidant/Oxidative Stress
|
|
TXNRD1
|
thioredoxinreductase 1
|
1.96
|
0.39
|
3.80
|
n/a
|
Anti-oxidant/Oxidative Stress
|
|
VCAN
|
|
-2.07
|
-0.41
|
n/a
|
n/a
|
This gene is involved in cell adhesion, proliferation, proliferation, migration and angiogenesis
|
Table1.Gene-Expression Data
A novel compound, Retinaldehyde γ-Cyclodextrin Complex (RCC), has been developed to open retinaldehyde to wider applications in the marketplace [12]. RCC and related compounds are prepared by the reaction of a polyene aldehyde, such as retinal, with a cyclodextrin, such as γ-cyclodextrin [13].
Materials and methods
Chemical structure study
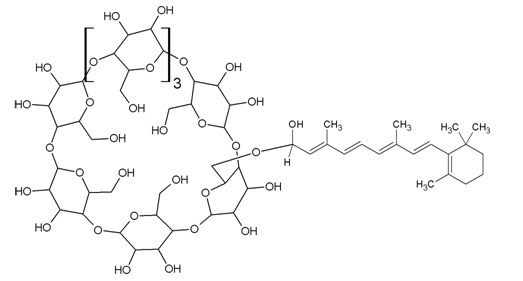
Structure 1: Retinaldehyde γ-cyclodextrin complex (RCC).
The mass spectra (ms) of RCC, purified via hplc, do not show a molecular ion. However, fragments at m/e 279, 285, 297, 299, and 371 arise from the fragmentation proposed in Figure 1. These fragments are not present in ms spectra of either γ-cyclodextrin or retinaldehyde.
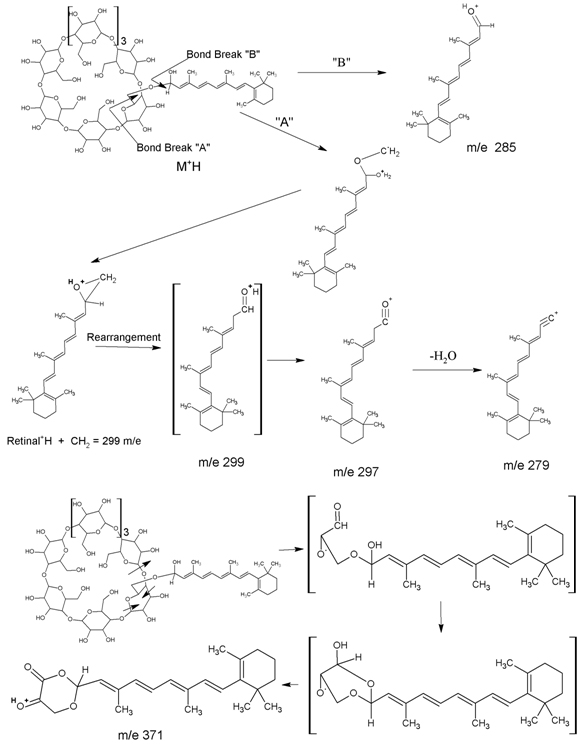
Figure 1. Mass Spectral Fragmentation of Retinaldehyde γ-Cyclodextrin Complex (RCC).
Gene expression study
The objective of the study was to understand how a topical material influences gene expression in the skin. The current study [14] was conducted using a full thickness in vitro skin culture model (MatTek, Epiderm EFT-400). Two test materials, RCC powder, and a 99% pure form of crystalline retinaldehyde (retinal), were each diluted in 100% dimethylsulfoxide (DMSO) to a final retinaldehyde concentration of 0.1%. A 15-μL volume of each test material was applied to the surface of each test culture. Cultures treated with 15 μL of 100% DMSO served as the control group. Cultures were collected 24 hours post-application for gene expression analysis.
Gene expression was analyzed using validated Taqman gene expression assays in the Taqman Low Density Array (TLDA) format. Analysis was carried out using the Genemarkers Standard Skin Panel, which contains assays for 92 target genes and four endogenous control genes. One additional target gene, HSP47, was assayed using a 96-well format. All genes in both formats were assayed in duplicate. A summary of the genes with statistically significant FC values is shown in Table 1. Negative values indicate decreased gene expression (down-regulation) and positive values indicate increased gene expression (up-regulation). RNA yield and quality was ensured using qPCR metrics based on up/down regulation of genes. NA=fold-change values were not greater than or equal to 2.0. Changes in gene expression greater than or equal to 2.0 are reported as linear fold change differences between test material and control groups (paired t-tests, p ≤ 0.05, N=4).
Exceptional Results
The gene-expression data (Table 1 ) clearly indicate RCC provides a topical method of treating, reducing the occurrence of, or improving the symptoms associated with melanogenesis, oxidative damage, inflammation, skin irritation from inflammation, loss of cell adhesion, loss of desquamation, extra-cellular including connective tissue matrix breakdown and skin tone loss thereof, loss of keratinization, cellular senescence, skin aging from cellular senescence, loss of skin whiteness, loss of skin barrier function, loss of skin firmness, inflammation from rosacea, skin disfigurements and skin discoloration from rosacea, inflammation from acne, skin wrinkles and fine lines from cellular senescence, cellular oxidation, loss of skin collagen, and topical wounds.
2021 Copyright OAT. All rights reserv
Conclusion
Retinaldehyde γ-Cyclodextrin Complex (RCC) now provides marketing opportunities for high-performance cosmetics that wish to utilize retinaldehyde into their line-up with unprecedented skin care attributes.
References
- Mukherjee S, Date A, Patravale V, Korting HC, Roeder A, et al. (2006) Retinoids in the treatment of skin aging: an overview of clinical efficacy and safety. Clin Interv Aging 1: 327-348. [Crossref]
- Merkviladze N, Gaidamashvili T, Tushurashvili P, Ekaladze E, Jojua N (2010) The efficacy of topical drugs in treatment of noninflammatory acne vulgaris. Georgian Med News 186: 46-50. [Crossref].
- Thielitz A, Abdel-Naser MB, Fluhr JW, Zouboulis CC, Gollnick H (2010) Topical retinoids in acne-an evidence-based overview. J Dtsch Dermatol Ges 11:S15-23. [Crossref]
- Darlenski R, Surber C, Fluhr JW (2010) Topical retinoids in the management of photodamaged skin: from theory to evidence-based practical approach. Br J Dermatol 163: 1157-1165. [Crossref]
- Gao SQ, Maeda T, Okano K, Palczewski K (2012) A microparticle/hydrogel combination drug-delivery system for sustained release of retinoids. Invest Ophthalmol Vis Sci 53: 6314-6323. [Crossref]
- Evans JE, McCaffery P (2010) HPLC/MS(N) analysis of retinoids. Methods Mol Biol 652: 149-162. [Crossref]
- Kane MA, Napoli JL (2010) Quantification of endogenous retinoids. Methods Mol Biol 652: 1-54. [Crossref]
- Kane MA, Folias AE, Napoli JL (2008) HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues, Anal Biochem 378: 71-79. [Crossref]
- Viguera AR, Villa MJ, Goñi FM (1990) A water-soluble polylysine-retinaldehyde Schiff base. Stability in aqueous and nonaqueous environments. J Biol Chem 265: 2527-2532. [Crossref]
- Cordero A, Leon-Dorantes G, Pons-Guiraud A, Di Pietro A, Asensi SV, et al. (2011) Retinaldehyde/hyaluronic acid fragments: a synergistic association for the management of skin aging. J Cosmet Dermatol 10: 110-117. [Crossref]
- Jomard et al., US Pat., 9,186,358 (November 17, 2015).
- Peter et al., Topical Therapies Based on Gene-Expression of Novel Retinaldehyde
- Cyclodextrin Complexes, J Dermatolog Clin Res 3(2): 1047 (2015); Peter et al., US Pat. 8,586,730 (November 19, 2013).
- Peter et al., US Pat. 8,410,079 (April 2, 2013); 8,859,269 (October 29, 2013).
- Genemarkers LLC, Project Report 024-022, June 6, 2013.


