Abstract
Background: In proteomics, the two-dimensional gel electrophoresis (2-D PAGE) technique is used to study the proteome which is defined as the whole protein supplement expressed by the genome of a living organism or a cell type at a given time, under defined circumstances, including post translation modifications. There are several published staining methods available that can be employed. The differences in the methods varies from sensitivity, cost, ease of use and subsequent application. There is sometimes a dilemma in which IPG range to choose differentially as there are various pH ranges on the market as well as the method of staining to employ when conducting 2D-PAGE. The study aim was to find the most appropriate IPG pH range and staining method for the identification of differently expressed proteins relating to meat tenderness in Nguni breed. Proteins were extracted from snap-frozen muscle (200 mg) homogenised in 1 ml TES buffer using Precellys homogeniser and quantified by a commercial kit at 750 nm (RC-DC Protein Assay), based on Lowry assay. Two isoelectric point ranges spanning the pH region 3-10 and 5–8, four staining methods: silver, Coomassie Brilliant Blue R250 and G250 as well as cyanide CyDyes were evaluated. The results show that the IPG pH 5-8 was more suitable than the pH 3-10. The Cy5 showed to be more sensitive and produced more visual spots than the other three methods. Due to practical factors such as cost and the inability to pick Cy5 spots manually, the Coomassie Brilliant Blue G250 staining method was chosen as the preferred one for future work.
Key words
2D gel electrophoresis, Nguni breed, gel staining methods, immobilized pH gradient, Coomassie Brilliant Blue G250
Abbreviations
2-D PAGE: Two-dimensional gel electrophoresis; SDS PAGE: sodium dodecyl sulphate polyacrylamide gel electrophoresis; IEF: isoelectric focusing; IPG: Immobilized pH gradient; ARC-API: Agricultural Research Council- Animal Production Institute; RMRDSA: The Red Meat Research and Development of South Africa; THRIP: Technology and Human Resources for Industry Programme
Introduction
One of the protein study disciplines in biology is the field of proteomics. Proteomics is the study of proteome, described as the whole protein supplement expressed by the genome of a living organism or a cell type at a given time, under defined circumstances, including post translation modifications [1]. Proteomics involves several techniques, one of which are two-dimensional gel electrophoresis (2-D PAGE). As pioneered by Klose [2] and O’Farrell [3] and improved by Görg et al [4], the technique is comprised of five stages i.e. isoelectric focusing (IEF) in the first dimension, sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) in the second dimension, staining, imaging and image analysis. Isoelectric focusing separates proteins based on their net charge and SDS-PAGE separates proteins based on molecular mass of protein subunits. Immobilized pH gradient (IPG) strips assist protein separation in the IEF dimension. The other role player in accomplishing success is the staining methods. There are several published staining methods available that can be employed. The differences in the methods varies from sensitivity, cost, ease of use and subsequent application. There is sometimes a dilemma in which IPG range to choose differentially as there are various pH ranges on the market as well as the method of staining to employ when conducting 2D-PAGE. The following are the most common used stains in 2D-PAGE i.e. Coomassie Brilliant Blue R250 and G250, silver, and SYPRO Ruby deep purple [5]. Apart from these commonly used stains, there are also methods such as minimal cyanide CyDyes labelling and iTRAQ that are gaining momentum [6,7]. While all these methods are commonly used they still have advantages and disadvantages such as mass spectrometry and specialised imaging equipment compatibility. For instance, silver staining Mortz et al. [8] and minimal CyDyes are not easily compatible with mass spectrometry. Although there are various staining methods there is no one method that surpasses the other and this is still a challenge to choose the best method for the specific application. In this study, two IPG pH ranges 3-10 and 5-8 were tested and four staining methods: silver, Coomassie Brilliant Blue R250 and G250 as well as CyDyes were evaluated with the aim to find the most appropriate IPG pH range and staining method for the identification of differently differentially expressed proteins relating to meat tenderness in the South African indigenous Nguni breed. Nguni cattle are well adapted animals and are tolerant of extreme warm and cold temperatures. They have shown to be resistance to internal and external parasites with excellent immunity to tick borne diseases. Nguni cattle are recognised to be an exceptional beef breed for optimum production under harsh African conditions.
Materials and methods
Animals and sampling procedures
Meat samples were collected from beef carcasses acquired from ARC-API abattoir Irene South Africa. The animals were finished off on a feedlot diet for a period of between 90-110 days at the ARC-API feedlot and were slaughtered at the ARC-API abattoir when they reached a live weight which would produce a carcass of Class A (no permanent incisors), and fat class 2 to 3 (1-≤5 mm) (South African Beef Classification System). Meat samples were sampled at longissimus lumborum one hour after slaughter and snap-frozen in liquid nitrogen and stored at -80°C.
Protein extraction
The frozen muscle (200 mg) was homogenised in 1 ml TES buffer as formulated according to Jia et al. [9]. A Precellys homogeniser (Bertin, le Bretonneux, France) was used at 5500 rpm, 2 × 20 sec,10 sec pause. After homogenising, the contents were centrifuged (30 min, 13000 rpm, 4 °C) to remove TES insoluble proteins. Protein concentrations were measured with a commercial kit at 750 nm (RC-DC Protein Assay, Bio-Rad, USA) based on Lowry assay in an ELX Universal micro-plate reader at 750 nm with BSA as a standard.
Two–dimensional gel electrophoresis
Protein separation in the first dimension was performed on an immobilised pH gradient IPG strips (Bio-Rad, USA), 24 cm, spanning the pH region 3-10 and 5-8. Protein extracts were loaded onto each IPG strip by passive rehydration for overnight at room temperature. For gels that were going to be stained with silver and coomassie Brilliant Blue stains 850 μg of protein was loaded. For the strips which were going to be stained with CyDyes, 50 μg of protein extract was loaded. Isoelectric focusing (IEF) was performed using the Ettan IPGPhor II unit (GE Healthcare BioSciences, Uppsala, Sweden) by means of a stepwise programme described as follows: 500 V for 2 h, increase to 1000 V for 2 h, increase to 10000 V for 3 h, 10000 V for 7:36 h. The second dimension proteins were separated on 12% SDS-PAGE using the Ettan DALT six large format vertical system (GE Healthcare Bio-Sciences) where after staining procedures followed i.e., Coomassie Brilliant Blue (G250 and R250), Silver and CyDyes (Cy5).
The Coomassie Brilliant Blue G250 stain was prepared as follows: 100 g of ammonium sulphate was dissolved into 650 ml of double distilled water, a 1 g of Coomassie Brilliant Blue G250 was added and stirred until Coomassie Brilliant Blue had dissolved. A 30 ml of o-phosphoric acid was added to the mixture followed by 200 ml of 99% ethanol and followed by double distilled water to make up 1000 ml. Gels were fixed for 60 minutes in fixing solution (o-phosphoric acid 85% 1.3% w/v and methanol 20% v/v) and stained for overnight. After staining, gels were transferred in neutralisation buffer (Tris-base 0.1 M, pH 6.5 o-phosphoric acid) for 1-3 minutes, and washed with 25% methanol for 1 minute and stored in stabilising solution (20% ammonium sulphate). The Coomassie Brilliant Blue R250 was prepared by weighing 0.25 g Coomassie Brilliant Blue R250 and dissolved in 1000 ml of double distilled water and heated up to 90°C. The gels were stained in a stainless steel tray and shaken for 30 minutes. Silver staining was performed according to Shevchenko et al. [10] except that silver nitrate was not chilled at 4°C, instead it was used at room temperature and formaldehyde was from ACE. The CyDye was prepared by following the labelling instruction included in the package (Amersham CyDye DIGE Fluors, minimal dyes for Ettan DIGE product booklet page 15-16). Gels were imaged and processed using Chemi-Doc Mp (Bio-Rad Hercules, CA, USA) equipped with Image Lab software.
Results and discussion
The images obtained (Figure 1) shows that there were no problems regarding passive overnight hydration of both pI 3-10 and 5-8. The results in Figure 1 show that with the pI 3-10 the proteins are focused and assembled mostly between pI 5-7. This focusing of the proteins around pI 5-7 led to investigation using IPG strip pI 5-8. Though some studies such as Czegledi et al. [11] used IPG strip pI 3-10 on the muscle tissue and found that proteins are evenly dispersed, for this study it was not the case. These maybe because of the type of the protein extract, different isoforms of protein and the environment which animals were raised. The results of using pI 5-8 are illustrated in Figure 2. The results illustrate four images from four different stains namely Coomassie Brilliant Blue G250 (A), Silver (B), Coomassie Brilliant Blue R250 (C) and CyDye 5 (D). There was not much difference between the two images A and C stained with two forms of Coomassie Brilliant Blue G250 and R250 as protein visualisation was the same, except that image C Coomassie Brilliant Blue R250 had a darker background which is not desired as compared to image A (Coomassie Brilliant Blue G250).
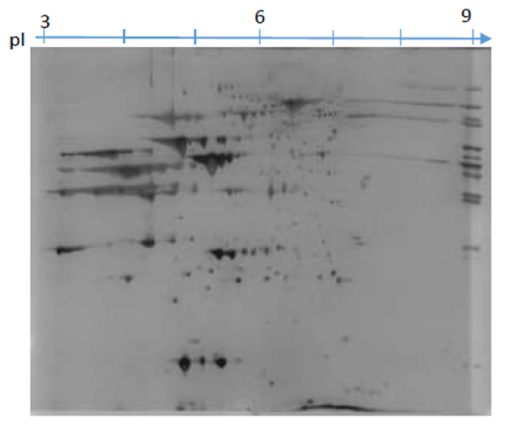
Figure 1. The gel images showing 2D-PAGE protein patterns of 850 μg of total TES protein extracts focused on the pI 3-10 stained with Coomassie G250.
There were major protein pattern differences in Figure 2 occurred between Coomassie Brilliant Blue G250 (A), silver (B) and Cy5 (D) with Cy5 displaying many protein spots followed by silver and Coomassie Brilliant Blue G250. In the image D (Cy5), there are more distinct protein spots and they are evenly dispersed through pI 5-8. This trend was found to be consistent with those reviewed in Chevalier [12] where by fluorescent dye (SYPRO Ruby) was found to be superior than Coomassie Brilliant Blue G250 and silver. There was a slight difference between image A (Coomassie Brilliant Blue G250) and B (silver). In B (silver) protein spots are visually sharper than those in A (Coomassie Brilliant Blue G250) but overall there was no much differences. Even though Czegledi et al. [11] concluded that sensitivity of silver was higher than that of Coomassie Brilliant Blue G250, in our study the sensitivities were similar with minor differences.
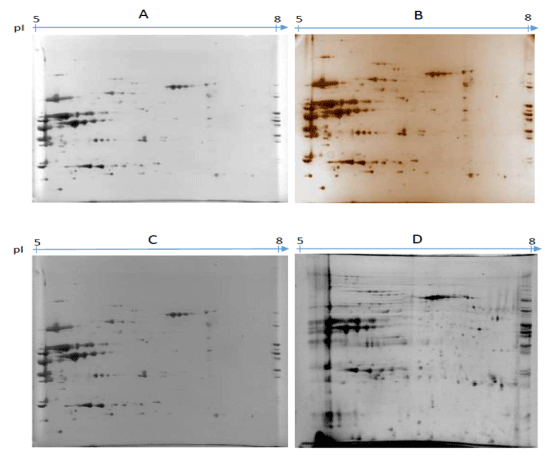
Figure 2. Major protein pattern differences.
Conclusion
The pI 5-8 IPG strips was chosen for the first dimension immobilised pH gradient due to most proteins being focused around the pI 5-7 region in the initial investigation using pI 3-10. Although fluorescent staining (Cy5) showed to be most superior to all the stains tested by showing more protein spots, Coomassie Brilliant Blue G250 stain was chosen as stain of interest as compared to silver and CyDyes because we were satisfied with the spot visualisation for the purposes of our studies and its advantages. The advantages of Coomassie Brilliant Blue G250 stain are the lower cost, visual inspection, easier operation, more convenient scanning procedure for image acquisition, better suitability for quantitative analysis than silver staining, and the capacity to allow possible modifications for fast or highly sensitive staining. Whereas CyDyes are difficult to visualise without an imager, it is challenging to pick protein spots manually and it is much more expensive. Silver was not chosen because the protocols show that silver staining is not compatible with mass spectrometry and its staining consistency was unreliable.
Ethics approval and consent to participate
2021 Copyright OAT. All rights reserv
The study was approved by the ARC_API ethics committee. The approval accession number ARC AEC-I 2010 001
Consent for publication
None
Competing interests
The authors declare that they have no competing interests
Funding
The Red Meat Research and Development of South Africa (RMRDSA) and the Technology and Human Resources for Industry Programme (THRIP) of the Department of Trade and Industry, South Africa funded this project.
Authors' contributions
KW carried the laboratory analysis and writing of the article, L reviewed the article, T helped in the laboratory during gel electrophoresis experiments, PE helped in the guidance of the article, G helped in the support and participated in the design of the study.
Acknowledgements
We would like acknowledge the Agricultural Research Council (ARC) for facilities and financial support. The Red Meat Research and Development of South Africa (RMRDSA) and the Technology and Human Resources for Industry Programme (THRIP) of the Department of Trade and Industry, South Africa for the funding of this project. We thank the Agricultural Research Council (ARC) Biochemistry team for technical support.
References
- Altelaar AF, Munoz J, Heck AJ (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet 14: 35-48. [Crossref]
- Klose, J (1997) Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26: 231-43.
- O'Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250: 4007-4021. [Crossref]
- Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, et al. (2000) The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21: 1037-1053. [Crossref]
- Panfoli I, Calzia D, Santucci L, Ravera S, Bruschi M, et al. (2012) A blue dive: from 'blue fingers' to 'blue silver'. A comparative overview of staining methods for in-gel proteomics. Expert Rev Proteomics 9: 627-634. [Crossref]
- Zieske LR (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot 57: 1501-1508. [Crossref]
- Hollung K, Veiseth E, Jia X, Færgestad EM, Hildrum KI (2007) Application of proteomics to understand the molecular mechanisms behind meat quality. Meat Sci 77: 97-104. [Crossref]
- Mortz, E, Krogh, T, N, Vorum, H, Görg, A (2000) Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1: 1359-1363
- Jia, X, Ekman, M, Grove, H, Færgestad, E, M Aass, et al. (2007) Proteome changes in bovine longissimus thoracis muscle during the early postmortem storage period. Journal of Proteome Research 6: 2720–2731.
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850-858. [Crossref]
- Czegledi L, Gulyas G, Radacsi A, Kusza S, Bekefi J, et al. (2010) Sample Preparation and Staining Methods for Two-Dimensional Polyacrylamide Gel Electrophoresis of Proteins from Animal Tissues. Animal Science and Biotechnologies 267–270.
- Chevalier F (2010) Standard dyes for total protein staining in gel-based proteomic analysis. Materials 4784–4792.


