Abstract
Bladder cancer causes an estimated 150,000 deaths per year worldwide. However, no major improvements in clinical outcomes have been achieved in the past several decades; therefore, a promising therapeutic agent is required. Recent studies revealed the existence of multiple subtypes of bladder cancer with distinct molecular signatures. To create novel therapeutics for such heterogeneous cancers, a target molecule should regulate various cancer-related signaling pathways. Here, we focused on the oncogenic miR-130 family (miR-130b, miR-301a, and miR-301b) as a novel therapeutic target for bladder cancer. The pharmacological inhibition of miR-130 family molecules by seed-targeting with an 8-mer tiny locked nucleic acid (LNA) inhibited 5637 bladder cancer cell growth, migration, and invasion by repressing stress fiber formation. Moreover, the miR-130-targeted LNA suppressed the phosphorylation of both FAK and Akt, resulting in the upregulation of two protein phosphatases, phosphatase and tensin homolog and protein tyrosine phosphatase, non-receptor type 11. In addition, administration of miR-130 family-targeted LNA significantly suppressed tumor growth in an in vivo bladder cancer xenograft model. Taken together, the miR-130 family-targeted LNA is expected to be a promising therapeutic agent for bladder cancer.
Key words
miR-130 family, locked nucleic acid, bladder cancer, PTEN, PTPN11
Abbreviations
miR: micro RNA; LNA: locked nucleic acid; FAK: focal adhesion kinase; PTEN: phosphatase and tensin homolog deleted from chromosome 10; PTPN11: Tyrosine-protein phosphatase non-receptor type 11; UTR: untranslated region
Introduction
Bladder carcinoma is the most common malignancy of the urinary tract worldwide, resulting in 150,000 deaths annually [1]. Recent genome-wide studies have revealed the existence of multiple molecular subtypes in bladder cancer [2–4]. Sjödahl et al. defined five major subtypes by mRNA expression analysis using bladder cancer specimens of all grades and stages. Among these subtypes, although both urobasal B and squamous cell carcinoma-like have the same poor clinical outcomes, their gene expression profile are quite different. Various molecular subtypes indicate that bladder cancer is highly heterogeneous. Therefore, therapeutic targets for bladder cancer should include genes that regulate broad cancer-related signaling pathways.
MicroRNAs (miRNA) are small non-coding RNA molecules that regulate gene expression through post-translational repression or mRNA degradation. In silico genome-wide analyses have shown that over 60% of all mammalian protein-coding genes are regulated by miRNAs [5,6]. Oncogenic miRNAs can therefore regulate broad tumor-related signaling pathways and have gained attention as therapeutic targets for cancer treatment [7–9]. Several studies have shown that the miRNA family contains a common seed sequence with functional overlap [10,11] and that the inhibition of miRNA family members significantly affects tumor progression. The miR-130 family (miR-130b, miR-301a and miR-301b), have also been shown to contribute cancer progression [12–14]. Furthermore, we recently found that the miR-130 family has tumor-promoting effects in bladder cancer [15]. Therefore, we examined the efficacy of the pharmacological inhibition of the miR-130 family by seed-targeting locked nucleic acid (LNA) to bladder cancer cells.
We report here that the miR-130 family-targeted LNA suppressed the phenotypes of bladder cancer cells both in vitro and in vivo, indicating that the miR-130 family-targeted LNA is expected to be a promising therapeutic drug for bladder cancer.
Materials and methods
Plasmid construction
To construct reporter plasmids for the miR-130 family (miR-130b, miR-301a, and miR-301b), PTEN, and PTPN11, the following oligonucleotides were used as primers:
hsa-miR-130b; sense 5¢-CTAGCGGCCGCTAGTATGCCCTTTCATCATTGCACTGG-3¢,
antisense 5¢-TCGACCAGTGCAATGATGAAAGGGCATACTAGCGGCCGCTAGAGCT-3¢, hsa-miR-301a; sense 5¢-CTAGCGGCCGCTAGTGCTTTGACAATACTATTGCACTGG-3¢, antisense 5¢-TCGACCAGTGCAATAGTATTGTCAAAGCACTAGCGGCCGCTAGAGCT-3¢, hsa-miR-301b; sense 5¢-CTAGCGGCCGCTAGTGCTTTGACAATATCATTGCACTGG-3¢, antisense 5¢-TCGACCAGTGCAATGATATTGTCAAAGCACTAGCGGCCGCTAGAGCT-3¢,
Sac1-PTEN 3 UTR; sense 5¢-GCGAGCTCGCTAAAGAGCTTTGTGATAT-3¢,
Sal1-PTEN 3'-UTR antisense 5¢-GCGTCGACATGCCATTTTTCCATTTCCA-3¢, Sac1-PTPN11 3¢-UTR; sense 5¢-TTGTGAGCTCTATTTTGCAGATTATGGGGA-3¢, Sal1-PTPN11 3¢-UTR; antisense 5¢-TTGTGTCGACCATTTGGCGACCAAAAACAC-3¢. The annealed fragments or PCR products were digested with SacI and SalI and inserted into the pmirGLO dual-luciferase miRNA target expression vector (Promega, Madison, WI, USA). The 3¢-UTRs of PTEN and PTPN11 were cloned from cDNA of 5637 cells using KOD-FX (Toyobo, Osaka, Japan).
Cell culture and transfection
The human uroepithelium cell line SV-HUC-1 and human bladder cancer cell line 5637 were cultured in F12K medium (Wako, Osaka, Japan) and RPMI 1640 medium (Wako), respectively, supplemented with 10% heat-inactivated fetal bovine serum and 100 mg/L kanamycin at 37°C under a 5% CO2 atmosphere.
The miR-130 family-targeted LNA (UCACGUUA) and control LNA (TCATACTA) were synthesized by GeneDesign (Osaka, Japan). These LNAs were transfected at a concentration of 50 nM using Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions.
Water-soluble tetrazolium salt-1 (WST-1) cell growth assay
Cell growth was examined in a WST-1 cell growth assay. The miR-130 family-targeted LNA-transfected 5637 cells were reseeded in a 96-well plate (2 × 103 cells/well) at 24 h after transfection and incubated for the indicated times. After incubation for 1 h with WST-1 reagent (Dojindo, Osaka, Japan) at 37°C, the optical density was read at a wavelength of 450/630 nm (measurement/reference) using a microplate reader (Bio-Rad, Hercules, CA, USA).
Wound healing assay
5637 cells were transfected with miR-130 family-targeted LNA at 24 h after seeding in a 12-well plate (6 × 104 cells/well) and incubated for 72 h. A wound was created in the monolayer of 5637 cells at ~90% confluence using a sterile 1-mL pipette tip. Cell pictures were recorded at 0 and 12 h after wound creation using a fluorescence microscope (Olympus, Tokyo, Japan).
Cell invasion assay
Cell invasion was measured using a CIM-plate and xCELLigence system (Roche, Basel, Switzerland) according to the manufacturer protocol.
Western blotting analysis
The whole cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride (PVDF; Millipore, Billerica, MA, USA) membrane using a semidry transfer system (Bio-Rad). The membranes were probed with specific antibodies as indicated and then incubated with horseradish peroxidase-conjugated antibody against mouse or rabbit immunoglobulin (Cell Signaling Technology, Danvers, MA, USA), followed by detection with enhanced chemiluminescence western blotting detection reagents (GE Healthcare, Little Chalfont, UK). An ImageQuant LAS4000 mini system (GE Healthcare) was used to detect chemiluminescence.
The following antibodies were used for immunological analysis in this study: anti-FAK polyclonal (Sigma, St. Louis, MO, USA), anti-p-FAK576 (Sigma), anti-Akt (CST), anti-p-Akt473 (Cell Signaling Technology), anti-PTEN polyclonal (Cell Signaling Technology), anti-PTPN11 polyclonal (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), and anti-actin polyclonal antibody (Sigma).
Microscopic observations
For fluorescence microscopic observation, cultured cells were grown on a micro coverglass, fixed by incubating in 4% formaldehyde, and then permeabilized with blocking buffer containing 5% bovine serum albumin and 0.1% Triton X-100 in phosphate-buffered saline. The permeabilized cells were incubated with primary antibody at 4°C overnight, followed by fluorochrome-conjugated secondary antibody for 1 h at room temperature. For F-actin staining, the permeabilized cells were incubated with 40 nM Acti-stain 488 Fluorescent Phalloidin (Cytoskeleton, Inc., Denver, CO, USA) at room temperature for 3 h. Next, the coverslips were mounted on a slide glass using Dapi Fluoromount-G (SouthernBiotech, Birmingham, AL, USA). Fluorescence images were obtained using a DP70 fluorescence microscope (Olympus).
Dual-luciferase reporter assay
A pmirGLO dual-luciferase miRNA target expression vector was used for the 3¢-UTR luciferase reporter assay (Promega). 5637 cells were co-transfected with miR-130 family-treated LNA and reporter construct containing the predicted miR-130 family binding site in the 3¢-UTR of the target genes. After 24 h of transfection, a dual-luciferase reporter assay was performed according to the manufacturer’s protocol (Promega). Luciferase activity was determined using a luminometer (Turner Biosystems 20/20 luminometer; Promega).
In vivo xenograft model and miR-130 family-targeted LNA/atelocollagen complexes administration
5637 cells (1 ´ 107 cells/mouse) were transplanted subcutaneously into female 8-week-old BALB/c nu-nu mice. When tumor growth was observed, the tumor-bearing mice were divided into two treatment groups (n=5/group) with an equivalent tumor volume. Atelocollagen (Koken, Tokyo, Japan) was used as a delivery carrier, which is known as a very effective drug delivery system for small interfering RNA and LNA molecules into tumors in vivo [16,17]. Individual mice were injected with 200 µL atelocollagen containing 2 nmol of the miR-130 family-targeted LNA or negative control LNA (HPLC-purified in vivo-grade LNA). LNA/atelocollagen complexes were injected on days 1, 7, 14, and 21. Tumor volumes were measured twice per week and evaluated using the following formula: tumor volume [mm3] = (major axis [mm]) × (minor axis [mm])2 × 0.5. Data are presented as the mean ± S.D. of five independent experiments.
Statistics
The results were expressed as the mean ± S.D. Differences between values were statistically analyzed using a Student’s t-test or one-way ANOVA with Bonferroni post-hoc tests (GraphPad Prism 5.0, GraphPad Software, La Jolla, CA, USA). A p-value < 0.05 was considered statistically significant.
Results
miR-130 family-targeted LNA suppresse2021 Copyright OAT. All rights reserv
To inhibit the function of the miR-130 family (miR-130b, miR-301a, and miR-301b) by one nucleotide molecule, we designed an 8-mer miR-130 family-targeted LNA (UCACGUUA) with a sequence complementary to nucleotide positions 2–9 containing a common seed sequence of the miR-130 family (Figure 1A). The function of the miR-130-targeted LNA was investigated in the bladder cancer cell line 5637, which shows the highest expression of miR-130 family molecules among a normal urothelium cell line (SV-HUC-1) and six bladder cancer cell lines (RT4, UM-UC-2, T24, EJ-1, J82, and 5637) [15], by a dual-luciferase reporter assay containing each miR-130 family binding sequence. The miR-130-targeted LNA successfully suppressed luciferase activity by binding to the miR-130 family target sequence (Figure 1B). Therefore, we used this 8-mer seed-targeting LNA for further functional analyses in 5637 bladder cancer cells.
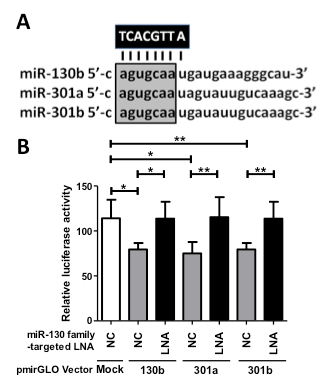
Figure 1. Design and functional verification of miR-130 family-targeted LNA in 5637 bladder cancer cells. (A); The miR-130 family-targeted LNA was designed with complementarity to nucleotide positions 2–9 of the miR-130 family sharing a common seed sequence (open box). (B); MiR-130 family reporter vectors containing a perfectly matched target site of miR-130b, miR-301a, or miR-301b (50 ng) were cotransfected with 50 nM miR-130 family-targeted LNA or negative control LNA into 5637 cells. Luciferase activity was determined using a dual reporter assay system. Data are presented as the mean ± S.D. of more than three independent experiments. *p < 0.05, **p < 0.01.
miR-130 family-targeted LNA suppresses growth, migration, and invasion of 5637 cells
To estimate the anti-cancer potential, we examined the effects of the miR-130 family-targeted LNA on cell growth, migration, and invasion in 5637 cells. Although we observed no effect on the cell growth of the normal urothelium cell line SV-HUC-1 (Figure 2A), the miR-130-targeted LNA significantly suppressed cell growth, migration, and invasion (Figure 2B–D) in 5637 cells. Since we found that miR-130 family molecules promote bladder cancer cell motility via stress-fiber formation and phosphorylation of focal adhesion kinase (FAK) at Tyr576 and Akt at Ser473 [15], we next examined the effect of miR-130-targeted LNA on stress fiber formation. Stress fiber formation in miR-130-targeted LNA tansfected-5637 cells was clearly suppressed compared to in control LNA-transfected cells (Figure 3A). Furthermore, we evaluated the effect of miR-130-targeted LNA on the phosphorylation of FAK and Akt. Both FAK and Akt phosphorylation levels were significantly decreased by miR-130-targeted LNA in 5637 cells (Figure 3B). Next, to identify the targets of miR-130-targeted LNA, we utilized target prediction programs (miRBase and microRNA.org), focusing on PTEN and PTPN11 as potential target genes. PTEN is a negative regulator of the PI3K/Akt signaling pathway [18,19], and both PTEN [20] and PTPN11 [21] can dephosphorylate tyrosine residues in FAK. We then confirmed whether PTEN and PTPN11 are target molecules of miR-130-targeted LNA in a dual-luciferase reporter assay using a reporter vector containing the 3¢-UTR region of either gene (Supplementary Figure S1). The luciferase activities of the PTEN of PTPN reporter vectors cotransfected with negative control LNA were significantly lower than that of the mock reporter vector alone. The activity was rescued by cotransfection with the miR-130-targeted LNA (Figure 3C). Immunoblot analyses showed that the transfection of miR-130-targeted LNA was increased by both PTEN and PTPN11 in 5637 cells (Figure 3D). These data suggest that miR-130 family-targeted LNA suppressed the phosphorylation of FAK and Akt and stress-fiber formation by restoring PTEN and PTPN11 expression that had been down-regulated by miR-130 family molecules.
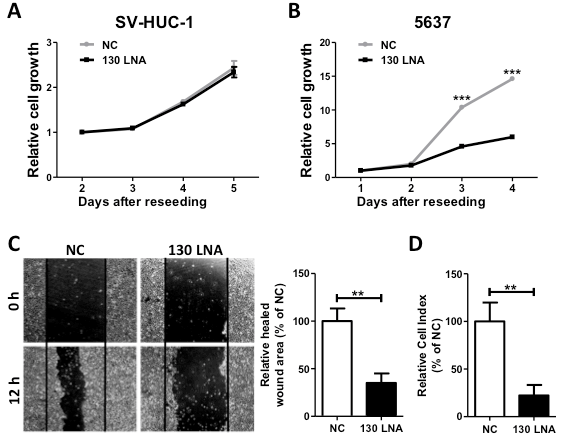
Figure 2. MiR-130 family-targeted LNA suppresses 5637 cell growth, migration, and invasion. The effect of miR-130 family-targeted LNA on SV-HUC-1 (A) or 5637 (B) cell growth was measured in a WST-1 assay. (C); Cell migration was determined using a wound healing assay. The wound was formed by scraping at 60 h after transfection and then relative cell migration was measured after 12 h. (D); The invasion assay was performed using the xCELLigence real-time cell monitoring system at 72 h after transfection. In all experiments, 50 nM miR-130 family-targeted LNA was transfected. Data are the mean ± S.D. of four (A), (B) nine, or (C) three (D) independent experiments. **p < 0.01; ***p < 0.001.
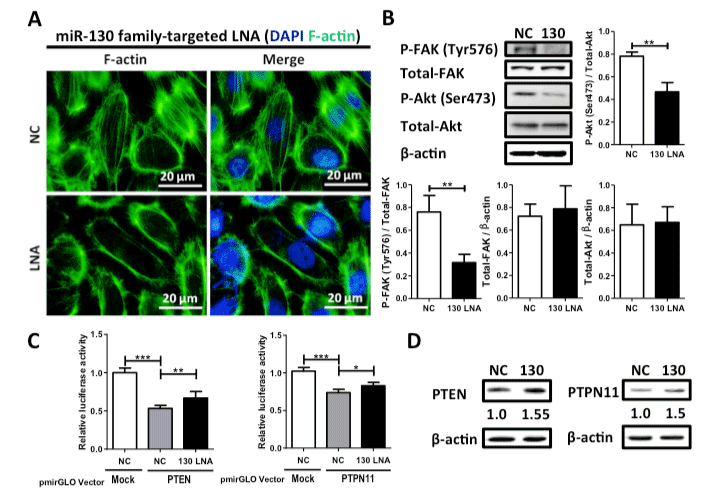
Figure 3. Effect of miR-130 family-targeted LNA on stress fiber formation and phosphorylation of FAK and Akt in 5637 cells. (A); Stress fiber formation was observed by F-actin staining with phalloidin. (B); Phosphorylation of FAK at Tyr576 and Akt at Ser473 was examined by western blot analysis. (C); A dual luciferase reporter assay was performed in 5637 cells. The cells were co-transfected with LNA and reporter plasmid containing the predicted miR-130 family binding site in the PTEN or PTPN11 3’-UTR. (D); Protein expression levels of both PTEN and PTPN11 were evaluated by western blot analysis. Data are the mean ± S.D. of three (B) or five (C) independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
miR-130 family-targeted LNA suppresses tumor growth in vivo
Finally, the anti-tumor activity of miR-130 family-targeted LNA was examined in a mouse xenograft model of 5637 cells. The increased tumor volumes following administration of control LNA were significantly suppressed by subcutaneous administration of miR-130-targeted LNA (Figure 4). The anti-tumor activity of miR-130-targeted LNA was determined based the decreased weight of removed tumors on day 27. These results indicate that miR-130 family-targeted LNA may be a novel nucleic acid therapeutic for bladder cancer.
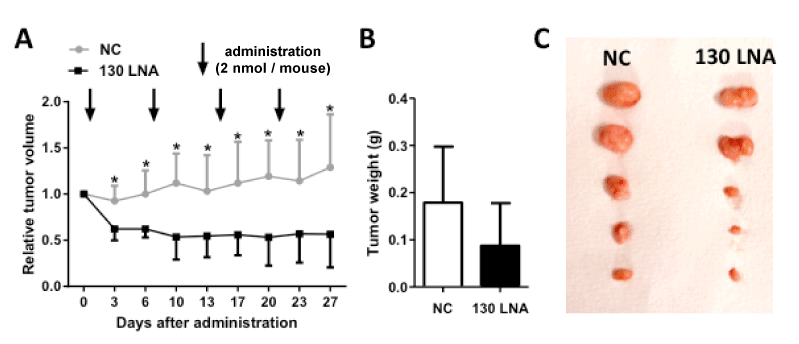
Figure 4. MiR-130 family-targeted LNA suppresses tumor growth in vivo. Control LNA and miR-130 family targeted LNA were administered at a dose of 2 nmol/mouse into 5637 cell-xenograft mice. (A); Relative tumor volume was calculated using the formula: tumor volume [mm3] = (major axis [mm]) × (minor axis [mm])2 × 0.5. Tumors resected on day 27 (B) were weighed (C). Data are mean ± S.D. (n = 5). *p < 0.05.
Discussion
In the present study, we designed 8-mer tiny LNA based on the seed sequence of the miR-130 family and demonstrated that the miR-130-tarageted LNA had inhibitory effects on the regulation of PTRN and PTPN11 by miR-130 family molecules. Moreover, the miR-130-targeted LNA was found to have significant anti-tumor activity.
PTEN functions as a tumor suppressor by negatively regulating the PI3K/Akt signaling pathway through lipid phosphatase activity in various types of cancers, including bladder cancer [19,20]. In bladder cancer, inactivation of PTEN is considered to be a trigger for the progression from a non-invasive to an invasive tumor [23,24], and urothelial-specific deletion of both p53 and pten causes invasive bladder cancer in mice [25]. PTEN acts as a dual-specificity protein phosphatase to regulate cell motility pathways in vitro [26]. We previously observed the inhibitory effect of PTEN overexpression on cell migration in 5637 cells (unpublished data); these results indicated that the potential for using PTEN against bladder cancer cell motility and subsequent tumor metastasis. Although the PTEN gene is mainly deleted by the loss of heterozygosity (LOH) in muscle-invasive bladder cancers [27], LOH in the PTEN region was detected in only 24.5% of bladder cancer specimens. Therefore, the presence of alternative mechanisms of PTEN regulation were predicted in bladder cancer, and the miR-130 family may act as an alternative upstream factor in PTEN expression.
In contrast to the PTEN, the physiological significance of PTPN11 in bladder cancer is not clearly understood. This is the first study suggesting that PTPN11 is a negative regulator of cell migration that suppresses FAK activity and stress fiber formation in bladder cancer cells. Functional inhibition of PTPN11 either by using a dominant-negative mutant [28] or pharmacological inhibitor [29] leads to the formation of stress fiber and focal adhesion. Furthermore, overexpression of FAK augments cell migration [30–32], and inhibition of the FAK-Paxilin signaling pathway decreases tumor metastasis in vivo [33]. Therefore, miR-130 family-targeted LNA may affect the metastasis of bladder cancer cells by regulating FAK signaling pathways via targeting PTEN and PTPN11 expression.
Compared to the 'classical' cytotoxic drugs, recently developed molecular-targeted drugs have shown many benefits in cancer patients while reducing side effects. However, over time, the cancer can become resistant to molecular-targeted drugs. To avoid this consequence, multi-targeted pharmaceuticals that can inhibit numerous cancer-related signaling pathways are required. In fact, administration of dasatinib, a multi-kinase inhibitor, showed better clinical outcomes for chronic myeloid leukemia patients compared to administration of imatinib. In advanced bladder cancer, a combination of a dual PI3K/mTOR inhibitor (PF-04691502) with a MEK inhibitor (PD-0325901) was tested in patient-derived xenograft models of bladder cancer, resulting in marked reduction of tumor growth [34]. Therefore, a multi-targeting strategy would be effective for treating cancer with a high degree of heterogeneity.
Considering these previous studies, miRNAs with various target genes are effective therapeutic targets for cancer treatment. Although a single miRNA typically targets 100–200 genes [35,36], the translational repression effect against each target gene is thought to be very weak. Therefore, targeting an miRNA family with overlapping functions in cancer enables wide and powerful inhibition of cancer-related signaling pathways [37]. Hullinger et al. showed that the complete inhibition of miR-15 family members using seed-targeting LNA induced more effective de-repression of target genes than the inhibition of each miRNA independently [38]. Similar results were observed for the miR-33 and miR-34 families [39,40]. Therefore, it is expected that targeting a seed region of the miRNA family by a tiny LNA will enhance functional outputs compared to the independent inhibition of a single miRNA.
Over the past two decades, no major breakthroughs have been achieved in the field of bladder cancer therapeutics. Therefore, novel drugs for bladder cancer are urgently required. This is the first report demonstrating the pharmacological inhibition of the miRNA family by seed-targeting LNA in bladder cancer. Although the largest amount of LNA accumulates in the kidney and liver [41], several studies have demonstrated effective LNA delivery into the orthotic bladder cancer via the urinary tract [42,43]. In conclusion, the miR-130 family-targeted LNA may be a promising nucleic acid therapeutic agent for bladder cancer treatment.
Conflicts of interest
K. Tsujikawa reports receiving a commercial research grant from Daiichi Sankyo, Fujifilm, and Tanabe Mitsubishi. No potential conflicts of interest were disclosed by the other authors.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893-2917. [Crossref]
- Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, et al. (2012) A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 18: 3377-3386.
- Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, et al. (2014) Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 111: 3110-3115. [Crossref]
- Choi W, Porten S, Kim S, Willis D, Plimack ER, et al. (2014) Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25: 152–165.
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-233. [Crossref]
- Friedman RC1, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92-105. [Crossref]
- Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, et al. (2014) MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 25: 469-483. [Crossref]
- Gao J, Li N, Dong Y, Li S, Xu L, et al. (2015) miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 34: 4142-4152. [Crossref]
- Ma Y, Zhang P, Wang F, Zhang H, Yang Y, et al. (2012) Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun 3: 1291.
- He L, He X, Lim LP, de Stanchina E, Xuan Z, et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130-1134. [Crossref]
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, et al. (2009) miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res 15: 5060–5072. [Crossref]
- Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, et al. (2010) miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 7: 694-707. [Crossref]
- Zhang W, Zhang T, Jin R, et al. (2014) MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J Exp Clin Cancer Res 33: 113. [Crossref]
- Funamizu N, Lacy CR, Parpart ST, Takai A, Hiyoshi Y, et al. (2014) MicroRNA-301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells. Int J Oncol 44: 725-734. [Crossref]
- Egawa H, Jingushi K, Hirono H, Ueda Y, Kitae K, et al. (2016) The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN, submitted.
- Tazawa H1, Tsuchiya N, Izumiya M, Nakagama H (2007) Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A 104: 15472-15477. [Crossref]
- Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, et al. (2013) miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS One 8: e79926. [Crossref]
- López-Knowles E, Hernández S, Malats N, Kogevinas M, Lloreta J, et al. (2006) PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res 66: 7401-7404. [Crossref]
- Tamguney T, Stokoe D (2007) New insights into PTEN. J Cell Sci 120: 4071-4079. [Crossref]
- Tamura M, Gu J, Danen EH, Takino T, Miyamoto S, et al. (1999) PTEN interactions with focal adhesion kinase and suppression of the extracellular matrix-dependent phosphatidylinositol 3-kinase/Akt cell survival pathway. J Biol Chem 274: 20693–20703. [Crossref]
- Tsutsumi R1, Takahashi A, Azuma T, Higashi H, Hatakeyama M (2006) Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol Cell Biol 26: 261-276. [Crossref]
- Hollander MC, Blumenthal GM, Dennis PA (2011) PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer 11: 289-301. [Crossref]
- Ho PL1, Kurtova A, Chan KS (2012) Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nat Rev Urol 9: 583-594. [Crossref]
- Knowles MA1, Hurst CD1 (2015) Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 15: 25-41. [Crossref]
- Puzio-Kuter AM1, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, et al. (2009) Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev 23: 675-680. [Crossref]
- Gildea JJ, Herlevsen M, Harding MA, Gulding KM, Moskaluk CA, et al. (2004) PTEN can inhibit in vitro organotypic and in vivo orthotopic invasion of human bladder cancer cells even in the absence of its lipid phosphatase activity. Oncogene 23: 6788–6797. [Crossref]
- Aveyard JS, Skilleter A, Habuchi T, Knowles MA (1999) Somatic mutation of PTEN in bladder carcinoma. Br J Cancer 80: 904-908. [Crossref]
- Kodama A, Matozaki T, Fukuhara A, Kikyo M, Ichihashi M, et al. (2000) Involvement of an SHP-2-Rho small G protein pathway in hepatocyte growth factor/scatter factor-induced cell scattering. Mol Biol Cell 11: 2565-2575. [Crossref]
- Schoenwaelder SM, Petch LA, Williamson D, Shen R, Feng GS, et al. (2000) The protein tyrosine phosphatase Shp-2 regulates RhoA activity. Curr Biol 10: 1523-1526. [Crossref]
- Cary LA, Chang JF, Guan JL (1996) Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J Cell Sci 109 : 1787-1794. [Crossref]
- Schlaepfer DD, Hunter T (1997) Focal Adhesion Kinase Overexpression Enhances Ras-dependent Integrin Signaling to ERK2/Mitogen-activated Protein Kinase through Interactions with and Activation of c-Src. J Biol Chem 272: 13189–13195. [Crossref]
- Gilmore AP1, Romer LH (1996) Inhibition of focal adhesion kinase (FAK) signaling in focal adhesions decreases cell motility and proliferation. Mol Biol Cell 7: 1209-1224. [Crossref]
- Lu YC, Chen CN, Wang B, Hsu WM, Chen ST, et al. (2011) Changes in tumor growth and metastatic capacities of J82 human bladder cancer cells suppressed by down-regulation of calreticulin expression. Am J Pathol 179: 1425-1433. [Crossref]
- Cirone P, Andresen CJ, Eswaraka JR, Lappin PB, Bagi CM (2014) Patient-derived xenografts reveal limits to PI3K/mTOR- and MEK-mediated inhibition of bladder cancer. Cancer Chemother Pharmacol 73: 525-538. [Crossref]
- Didiano D, Hobert O (2006) Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 13: 849-851. [Crossref]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259-269. [Crossref]
- van Rooij E, Kauppinen S2 (2014) Development of microRNA therapeutics is coming of age. EMBO Mol Med 6: 851-864. [Crossref]
- Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, et al. (2012) Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 110: 71-81. [Crossref]
- Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, et al. (2013) Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med 5: 212ra162. [Crossref]
- Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, et al. (2012) Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A 109: 17615-17620. [Crossref]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, et al. (2011) Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43: 371-378. [Crossref]
- Nogawa M, Yuasa T, Kimura S, Tanaka M, Kuroda J, et al. (2005) Intravesical administration of small interfering RNA targeting PLK-1 successfully prevents the growth of bladder cancer. J Clin Invest 115: 978-985. [Crossref]
- Shimada K, Fujii T, Tsujikawa K, Anai S, Fujimoto K, et al. (2012) ALKBH3 contributes to survival and angiogenesis of human urothelial carcinoma cells through NADPH oxidase and tweak/Fn14/VEGF signals. Clin Cancer Res 18: 5247-5255. [Crossref]




