Abstract
Nepal is geographically located between India and China, a region containing significant Tuberculosis (TB) and Multi-Drug Resistance (MDR-TB) burdens. However, limited information is available on the phylogenetic diversity of Mycobacterium tuberculosis (Mtb) in Nepal. To gain further insight into the diversity of Mtb in Nepal, consecutive clinical samples from 176 newly diagnosed pulmonary tuberculosis patients were collected from two hospitals in Nepal. Insertion Site IS6110 Fluorescent Amplified Fragment Length Polymorphism (FAFLP) PCR and rpoB sequence analysis were carried out on genomic DNA extracts of cultured strains to assign them to accepted genetic lineages and identify MDR-TB. In this study, the IS6110 based characterisation showed a prevalence of 36.36% Central Asian Strain (CAS), 18.75% Beijing, 7.95 % Haarlem, 3.97% X, 2.2% each of Latin American Mediterranean (LAM), T-Uganda and T, 1.7% S and 24.4% were unassigned. Further, 3.9% of total M. tuberculosis isolates were of rifampicin resistant genotypes thus indicating that the prevalence of MDR could be higher than the country wide prevalence of MDR among new TB cases (2.2%) as reported by the national drug resistance survey carried out in 2011/2012.
Keywords
Tuberculosis; IS6110; FAFLP; Rifampicin resistance; TB lineages
Introduction
TB is ranked as the sixth leading cause of death among the top 20 causes of death in Nepal. According to National Tuberculosis Control Programme (NTCP) in Nepal, in 2014 37,025 TB cases were registered and among them 15,947 (43%) cases were new sputum smear positive TB cases. It was estimated by WHO [1] that 4.6 (2.1-7.5) thousand people in Nepal died from TB in 2014. Even though short course TB drug treatment regimen could cure around 89% of cases, TB mortality was still unacceptably high in Nepal. Since 2006, the STOP TB strategy has been adopted by NTCP. However Drug resistant TB (DR-TB) still threatens national TB control and is a major public health concern. The proportion of MDR-TB cases in new cases was 2.2% and retreatment cases were 15.4%. Even though the Millennium Development Goal (MDG) to halt and reverse TB incidence has been achieved in all six WHO regions, work remains to be done to prevent the deaths from this dreadful disease [1].
The identification of the number and position of Insertion Sequence IS6110 elements in the Mtb genome has been widely used as a genomic tool for the rapid fingerprinting of isolates of Mycobacterium tuberculosis complex (MTBC) [2]. IS6110 based Restriction Fragment Length Polymorphism (RFLP) is considered as the ‘gold standard’ typing method for strains with more than five copies [3-5]. As IS6110 transposition is among the first genetic changes to occur in strains from a transmission chain [6], this marker has also been used for outbreak analysis [5].
Modification of the conventional IS6110 typing method, using differentially labelled primers has allowed characterisation of Mtb isolates into the key genetic lineages more rapidly than traditional methods [7]. This approach can be facilitated with automation, which enables this technique to be performed in a high throughput setting. The fragment patterns generated indicates both copy number and insertion site of IS6110 in the genome [8,9]. The patterns generated correlates directly with other independent markers and can be used for transmission investigation locally or internationally. Specific fragments are common in genetically related lineages and do not occur in other groups (e.g. spoligotype groups such as Beijing and the Euro-American lineage which contain the Latin American Mediterranean (LAM), Haarlem, S, T and X spoligotype groups). The patterns generated correlate directly with other independent markers and can be used for transmission investigation locally/internationally. Principal Genetic Groups (PGGs) can be assigned to Mtb strains based on the combination of polymorphism located at katG codon463 and gyrA codon95 in the respective genomes [10] or spoligotypes [11] or global phylogeny classification based on whole genome sequences [12].
Limited data are available on the characterisation of Mtb strains and genotypes circulating in Nepal. A key factor is the geographical location of Nepal, interlocked between China and India, two countries, which together account for approximately a third of annual global new cases (11% and 24%, respectively) [1]. A recent study of 261 Nepalese isolates found any drug resistance (any drug resistance has been defined as resistance to isoniazid, rifampicin, streptomycin, ethambutol, fluoroquinolones, and/or aminoglycosides) in 12.8%% of Mtb strains that were new untreated cases, with the most frequent lineages reported as CAS/Delhi (40.6%), East Asian (including Beijing) (32.2%), Euro-American (15.7%) and Indo-oceanic (11.5%) [13]. To gain further insight into the characteristics and diversity of Mtb in Nepal, our study aimed to categorise isolates for the first time using IS6110 FAFLP PCR and to assign them to different genetic lineages. Secondly, the level of MDR would be characterized in the population using rpoB (rpoB gene encodes for the Beta subunit of bacterial RNA polymerase) sequencing of the Rifampicin Resistant Determining Region (RRDR) as a predictive surrogate [14-17].
Methods
Strains
Sputum samples from 176 consecutive new TB patients over one year were collected between 2007 and 2008 and cultured alongside routine diagnostics from two Nepalese tuberculosis reference centres located in the Kathmandu valley: the National Tuberculosis Centre (NTC) and the German Nepal Tuberculosis Project (GENETUP). The patient population represented local and referred cases from across Nepal. Bacterial genomic DNA from isolated strains was extracted by the Cetyltrimethylammonium Bromide (CTAB) method [18] at the Mycobacterial Research Laboratories (MRL) in Anandaban Hospital. Informed consent was not required at the time of this study, as samples were collected with routine clinical care and all patient identifiers were anonymized; however, all patients were provided an explanation and were only included upon provision of verbal informed consent. Study procedures were reviewed and approved by NTC and GENETUP. The results for the drug sensitivity tests were unavailable during the entire duration of this study.
IS6110 FAFLP PCR, Fragment Sizing & Analysis
Genomic DNA was digested with the restriction enzymes MseI and TaqI followed by ligation with double stranded TaqI restriction site specific adaptors. The adaptor ligated DNA was amplified following previously published PCR conditions using four fluorescently labelled adaptor specific TaqI forward primers - 5’- CGATGAGTCCTGACCGA*/C*/T*/G* each labelled with a single unique selective nucleotide at the 3’ end and an IS6110 sequence specific reverse primer- 5’- CTGACATGACCCCATCCTTT [9]. In a total volume of 20 µl, 1 µl of the adaptor ligated DNA was added to the reaction containing 1X reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs (Invitrogen, UK) , 1 µM of labelled Taq I forward primer, 1 µM of IS6110 reverse primer and 1 U of recombinant Taq polymerase (Invitrogen, UK). The following PCR conditions were carried out in a Veriti thermocycler (Applied Biosystems, UK): 94°C for 15min followed by 35 cycles of 94°C for 20s, 66°C for 30s and 72°C for 2 min with the 66°C annealing temperature reducing by 1°C every cycle for nine cycles and the last 25 cycles at 56°C. Finally, an extension of 72°C for 60min was carried out before further manipulations. The fragments were separated on an ABI genetic analyser 3730XL (Applied Biosystems, UK), sized using PeakScanner v1.0 software (Applied Biosystems) and identified using their fluorescent tag (Figure 1). The four-dye FAFLP data collected from the different profiles were then recorded and compared with a reference collection of Mtb isolates [19] using BioNumerics software v6.1 (Applied Maths Inc., Belgium). Fragments common to different lineages (defined as being present in >50% of strains in a particular genetic lineage) were recorded for each Nepalese strain and compared with a fully characterised global collection as detailed by Thorne et al. [20]. These data were then used to build a dendrogram using the Dice coefficient of similarities to compare the similarity matrix and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) derived cluster analysis with cophenetic correlation for the branch quality.
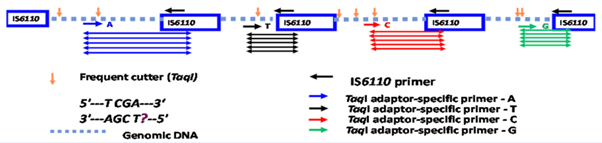
Figure 1: Pictorial representation of the IS6110 FAFLP PCR methodology. Coloured fragments are amplified using uniquely labelled adapter specific primers. For example, blue coloured fragments are amplified if the primers are labelled with 6-FAM fluorescein ‘blue’ dye, red coloured fragments with PET dye, green coloured fragments with VIC dye and black coloured fragments with NED dye.
rpoB Analysis
The 81 bp Rifampicin Resistant Determining Region (RRDR) of the rpoB gene of all strains were sequenced using published primers [21] and analysed in BIOEDIT software using ClustalW alignment parameters. The PCR was carried out in a total volume of 50µl where 1µl of the DNA was added to the reaction containing 1xPCR reaction buffer, 1.5mM MgCl2, 0.2mM dNTPs (Invitrogen, UK), 20µM each of both rpoB-RRDRforward (5’-CGATCACACCGCAGACGTTGA) and reverse primers (5’- GGCACGCTCACGTGACAGACC) and 5U recombinant Taq polymerase (Invitrogen, UK). The following PCR conditions were carried out using a Veriti thermocycler (Applied Biosystems, UK): 94˚C for 2 min followed by 35 cycles of 94˚C for 30 sec, 60˚C for 30 sec and 72˚C for 1 min. Finally, an extension of 72˚C for 10 min was performed before cleaning the products using AmpureXP magnetic beads (Beckman Coulter, UK) according to the manufacturer’s protocol and sequencing using the forward primer, rpoB-RRDR forward.
Results
Analysis of Data using BioNumerics software v6.1
Of the 176 DNA extracts from isolates analysed, the majority of the samples 97 (55.4%) belonged to either the spoligotype-defined Central Asian Strain (CAS) lineage (64 i.e. 36.6%) or the Beijing lineage (33 i.e. 18.8%) grouping under PGG1 and the rest of the samples group under either PGG2 (1.7% S, 3.97% X, 7.95% Haarlem and 2.27% LAM, 2.27% T-Uganda) or PGG3 (2.27% of T) (Table 1). Forty three samples (24.4%) grouped under “unassigned” group. Common fragments seen were exactly the same as the earlier published report by Thorne et al, (2011) except for an additional fragment, 78.4 G, for the CAS lineage. A dendrogram was generated using only the IS6110 FAFLP data (Figure 2) confirming again the above mentioned lineages in relation to the PGGs.
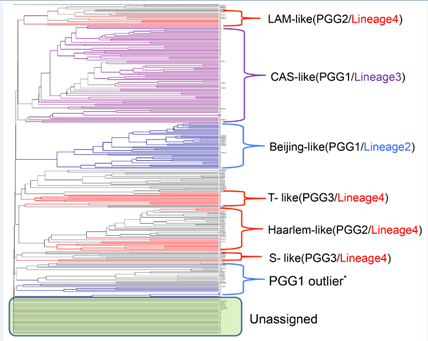
Figure 2: UPGMA derived dendrogram showing the predominant genetic lineages / spoligotypes of 176 Nepalese Mtb isolates.
Coloured branches represent Nepal samples following Gagneux’s global phylogeography of MTBC [12] and black branches represent the in-house global Mtb collection. PGG1 outliers share one IS6110 copy with the PGG1 group. Those which contained one IS6110 copy but could not be assigned to any group (unassigned group in the figure) are shown within the green box.
Table 1: Common fragments identified using IS6110 FAFLP PCR in TB genetic lineages between the 176 bacterial DNA isolates in Nepal.
|
PGG /spoligotype/ sub-lineage
|
Common fragment sizes
|
No. Of Nepal strains
|
|
PGG1-CAS
|
78.4G ,92.0B , 117.9R, 206.2G , 275.1R
|
64/176
|
|
PGG1-Beijing
|
101.7B, 102.5Y, 139.1R, 180.7Y, 254.8G, 332.4R, 353.5B
|
33/176
|
|
PGG2 –Haarlem
|
87.0Y, 89.4G, 148.7B, 300.2R (H/X), 445.7Y
|
14/ 176
|
|
PGG2-LAM
|
71.5Y, 105.2R, 116.1R
|
4/ 176
|
|
PGG2-S
|
88.0G, 112.9R, 217.2R, 445.3G
|
3/ 176
|
|
PGG2-X
|
83.8Y, 300.2R(H/X)
|
7/ 176
|
|
PGG2-T Uganda
|
88.9Y, 119.5G, 122.9Y, 228.4Y, 266.8R
|
4/ 176
|
|
PGG3- T
|
81.3R, 192.4R, 360R
|
4/176
|
|
Ungrouped
|
|
43/176
|
Where B-Blue coloured fragment R- Red coloured fragment G-Green coloured fragment and Y-Black/ Yellow coloured fragment seen in the electropherogram. PGG represents Principal Genetic groups according to Sreevatsan et al. [10], spoligotypes follow spolDB4 classification [4] and sub-lineages are grouped following Gagneux’s classification [8].
rpoB Analysis
Of 176 DNA extracts analysed for rpoB mutations, seven samples (3.9%) had a single non-synonymous base change which would likely confer resistance to rifampicin (Table 2). Six of these seven samples showed a second base mutation in a codon triplet whereas sample N70 showed a first base mutation.
Table 2: List of mutations seen in rpoB Rifampicin Resistance-Determining Region (RRDR) of rifampicin resistant M. tuberculosis isolates from Nepal.
|
Sample
|
Mutated locus
|
Nucleotide modification
|
Amino acid modification
|
FAFLP derived
|
- N70
|
516
|
GAC > TAC
|
(Asp>Tyr)
|
PGG1- Beijing group
|
- N10
|
522
|
TCG > TTG
|
(Ser>Leu)
|
PGG1- CAS group
|
- N25
|
526
|
CAC > CTC
|
(His>Leu)
|
PGG3 -like
|
- N63
|
526
|
CAC > CGC
|
(His>Arg)
|
PGG3-like
|
- N34
|
531
|
TCG > TGG
|
(Ser>Trp)
|
PGG3- T group (H37Rv-like)
|
- N46
|
531
|
TCG > TTG
|
(Ser>Leu)
|
PGG3 (H37Rv-like)
|
- N62
|
531
|
TCG > TTG
|
(Ser>Leu) 2021 Copyright OAT. All rights reserv
|
Ungrouped
|
* PGG represents Principal Genetic Group.
Discussion
It has been demonstrated previously that IS6110 FAFLP PCR can be used to delineate the phylogeny of MTBC as shared common fragments can determine the different lineages in a geographical location by comparison with a reference database collection [7]. As limited lineage information is available from strains in Nepal, we have applied the IS6110 method published recently on mapping the IS6110 sites in H37Rv [9] and also carried out rpoB sequencing to further characterize strains from this important region.
Fifty five percent of the 176 Nepalese strains analysed belong to the CAS (36.6%) and Beijing (18.8%) modern genetic spoligotypes (PGG1). The remaining 24.4 % of the samples belong to the PGG2 and PGG3 groups (Haarlem, LAM, S, X, T-Uganda and T). However, a limitation of this technique is its difficulty to characterise the samples with less than 4-5 copies of IS6110 as seen in the unassigned group (24.4%) in figure 2, which can be overcome by the use of other typing techniques like Mycobacterial Interspersed Repetitive Units- Variable Number Tandem Repeats (MIRU-VNTR) [20]. The geographical position of Nepal is likely to have influenced this distribution, with a mixture of predominantly Beijing lineage from the North of the Himalayas and the CAS lineage from the south [22]. The fairly high percentage of mainly European lineages (Haarlem, LAM and T, 12.5%) indicates that there has also been mixing of the different lineages over an extended time and that European travellers/migrants to South East Asia and Nepal may have transmitted European strains to the local population. According to Malla et al. [13], fifty strains had any drug resistance and sixteen (6.1%) out of 261 isolates were MDR. Among the fifty any drug resistant strains, 29 cases were previously treated and twenty-one were new untreated cases (8.04% of 261 total strains and 12.8% of 164 new untreated cases). In this study, MDR-TB was tested by using rifampicin as the resistance marker and 7 isolates out of 176 were found (3.9%) from new untreated cases possessing drug resistance genotypes. However, their reported MDR percentage was based on the total number of isolates, of which 37.2% of isolates were from previously treated cases, which may enhance a probability of drug resistance development compared to untreated cases. Our results indicate that the prevalence of Rifampicin (RIF) resistant TB (surrogate marker for MDR) was higher than the nationally reported 2.2% MDR in new untreated cases. Further, our results are concordant with a recently conducted study by Creswell et al. [23], where they have shown that the genotypic rifampicin resistance in newly diagnosed TB patients to be 3.3% in Nepal [23].
Conclusions
The IS6110 FAFLP data from our study reiterates the fact that the geographic location of Nepal is the key for the circulation of PGG1 TB lineages, CAS and Beijing, which were predominant in India and China respectively. Further the RRDR study correlates with the recent work by Creswell et al. showing that prevalence of MDR-TB may be marginally higher than the national average in new untreated TB cases. As the monitoring of TB is important in Nepal, this simple and informative PCR-based molecular epidemiological technique would prove useful for the study of outbreaks of the disease and also to detect cross-contamination between different strains or isolates in resource poor settings. The most common mutation site in the RRDR is at codon 531 and parallels the findings of earlier studies [21,22].
Authorship
All authors mentioned above gave substantial intellectual contribution to this manuscript.
Acknowledgments
We thank gratefully to the staff of the NTC and GENETUP from the Kathmandu valley in Nepal. We would like to thank all staffs from MRL, Anandaban Hospital in Lalitpur, Nepal for their technical help. We are also grateful to Dr. Ali Al-Shahib from Bioinformatics Department (PHE, UK) and Mr. Jon White from Medical Illustration Department (PHE, UK) for their technical expertise.
Funding Information
The authors received no specific funding for this work.
Competing Interest
The authors declare that they have no competing interests.
References
- WHO (2014) Global tuberculosis report 2014.
- Haas WH, Butler WR, Woodley CL, Crawford JT (1993) Mixed-Linker Polymerase Chain Reaction?: a New Method for Rapid Fingerprinting of Isolates of the Mycobacterium tuberculosis Complex. Tuberculosis 31: 1293-1298.
- Otal I, Martín C, Vincent-Lévy-Frebault V, Thierry D, Gicquel B (1991) Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol 29: 1252-1254.
- de Boer AS, Borgdorff MW, de Haas PE, Nagelkerke NJ, van Embden JD, et al. (1999) Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis 180: 1238-1244. [Crossref]
- van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, et al. (1993) Strain Identification of Mycobacterium tuberculosis by DNA Fingerprinting?: Recommendations for a Standardized Methodology. J Clin Microbiol 31: 406-409. [Crossref]
- Schürch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, et al. (2010) The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infection, genetics and evolution? Infect Genet Evol 10: 108-114. [Crossref]
- Thorne N, Borrell S, Evans J, Magee J, García de Viedma D, et al. (2011) IS6110-based global phylogeny of Mycobacterium tuberculosis. Infection, genetics and evolution. Infect Genet Evol 11: 132-138.
- Borrell S (2009) Comparison of four-colour IS6110-fAFLP with the classic IS6110-RFLP on the ability to detect recent transmission in the city of Barcelona, Spain. Tuberculosis (Edinburgh, Scotland) 89: 233-237.
- Moganeradj K, Abubakar I, McHugh TD, Sonnenberg P, Arnold C (2013) Insertion site mapping for repeated elements in Mycobacterium tuberculosis. J Microbiol Methods 92: 192-196. [Crossref]
- Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. (1997) Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA 94: 9869-9874. [Crossref]
- Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. (2006) Mycobacterium tuberculosis complex genetic diversity?: mining the fourth international spoligotyping database ( SpolDB4 ) for classification, population genetics and epidemiology. BMC Microbiol 6: 23. [Crossref]
- Gagneux S (2012) Host-pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol SciPhilosophical 367: 850-859. [Crossref]
- Malla B, Stucki D, Borrell S, Feldmann J, Maharjan B, et al. (2012) First insights into the phylogenetic diversity of Mycobacterium tuberculosis in Nepal. PLoS One 7: e52297. [Crossref]
- Luita Nice CO, Jairo da Silva MS, Luiz Alexandre VM, Sônia Cristina OM, Antonio M (2016) Detection of multidrug-resistant Mycobacterium tuberculosis strains isolated in Brazil using a multimarker genetic assay for katG and rpoB genes. Braz J inf dis 20: 166-172.
- Tripathi R, Sinha P, Kumari R, Chaubey P, Pandey A, et al. (2016) Detection of rifampicin resistance in tuberculosis by molecular methods: A report from Eastern Uttar Pradesh, India. Indian J Med Microbiol 34: 92-94. [Crossref]
- Suresh N, Singh UB, Arora J, Pande JN, Seth PSuresh, N, et al. (2006) Rapid detection of rifampicin-resistant Mycobacterium tuberculosis by in-house, reverse line blot assay. Diag Microbiol Infec Disease 56: 133-140.
- Somoskovi A, Parsons LM, Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2: 164-168.
- van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD (1991) Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29: 2578-2586. [Crossref]
- Kristin K, Catherine A, Angel C, Cristina MG, Walter HH (2005) Discriminatory Power and Reproducibility of Novel DNA Typing Methods for Mycobacterium tuberculosis Complex Strains Discriminatory Power and Reproducibility of Novel DNA Typing Methods for Mycobacterium tuberculosis Complex Strains. J Clin Microbiol 43: 5628-5638.
- Thorne N, Evans JT, Smith EG, Hawkey PM, Gharbia S, et al. (2007) An IS6110-targeting fluorescent amplified fragment length polymorphism alternative to IS6110 restriction fragment length polymorphism analysis for Mycobacterium tuberculosis DNA fingerprinting. Clin Microbiol Infect 13: 964-970. [Crossref]
- Arnold C, Westland L, Mowat G, Underwood A, Magee J, et al. (2005) Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin Microbiol Infect 11: 122-130. [Crossref]
- Ajay P, Chie N, Yukari F, Haruka S, Basu DP, et al. (2012) Molecular characterization of multidrug-resistant Mycobacterium tuberculosis isolated in Nepal. Antimicrob Agents Chemother 56: 2831-2836.
- Creswell J, Rai B, Wali R, Sudrungrot S, Adhikari LM, et al. (2015) Introducing new tuberculosis diagnostics: the impact of Xpert(®) MTB/RIF testing on case notifications in Nepal. Int J Tuberc Lung Dis 19: 545-551. [Crossref]


