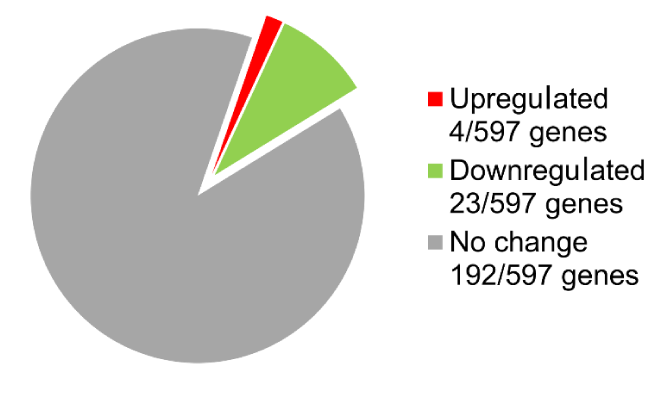Aim: OM-X® comprises edible bacterium-fermented plants and is used worldwide as a dietary supplement. The study aims to determine the biological effects of OM-X®.
Methods: OM-X® was added to cultured lower gastrointestinal tract cells from Wistar rats; Glucagon-like peptide-1(GLP-1) was quantified by enzyme-linked immunosorbent assay. RAW264.7 cells were cultured with OM-X® only or OM-X® plus Lipopolysaccharide (LPS); after 24 h, nitric oxide (NO) and cytokines production were measured in vitro. For in vivo investigations, 1% OM-X® was administered to ICR mice for 7 days, and 597 liver gene expressions were analyzed by focused DNA microarray.
Results: GLP-1 levels increased concentration dependently in rat lower gastrointestinal tract cells cultured with OM-X®. Similarly, OM-X® only or OM-X® plus LPS both increased RAW264.7 cell NO and cytokines (IL-6, and TNF-α) production activities. Mice administered OM-X® orally exhibited differential liver gene expression. Particularly, OM-X® significantly elevated the expressions of 4 genes and reduced the expressions of 23 kinds of inflammatory and apoptosis-associated genes.
Conclusions: OM-X® directly stimulated gastrointestinal tract cells and RAW264.7 cells to enhance GLP-1, NO, IL-6, and TNF-α production, possibly regulating blood glucose levels and stimulating an immune response. Modulation of liver gene expression showed that orally administrating OM-X® may have biological roles, including immunomodulatory and cytoprotective effects.
dietary supplement, GLP-1, NO, cytokines, DNA microarray
OM-X® is a dietary supplement developed by Dr. Iichiroh Ohhira using an original fermentation method in Japan. OM-X® is made from many kinds of vegetables, fruits, seaweeds, and mushrooms fermented using 12 strains of lactic acid bacteria (LAB) and bifidobacteria. These materials were obtained from the natural environment by strict selection from Japanese farms and hilly areas. After collecting the materials, they were mixed and fermented at room temperature for a few years in an exclusive clean factory of BIOBANK Co., Ltd., (Okayama, Japan). OM-X® is strictly made in compliance with the International Organization for Standardization (ISO) 9001. Nutritionally, OM-X® provides carbohydrate (oligosaccharides and dietary fiber), fats (short-chain fatty acids), proteins, amino acids, vitamins, and minerals [1] . OM-X® has been consumed in many countries, with no serious adverse reactions reported. Furthermore, its safety has been confirmed by tests in animals (mutagenicity and acute toxicity tests) and by a clinical trial in humans (phase I) [2].
Previously reported studies on OM-X® have revealed various functions, including improvement of leaky gut syndrome (LGS) in bacterium-infected model mice of LGS through proliferation of intestinal epithelial cells, anti-inflammatory action on Citrobacter rodentium-infected enteritis mice [3]. Human intervention studies have shown that OM-X® increased bone density of healthy male and female subjects and increased the maximum oxygen consumption (VO2 max) of athletes [4]. Improvement of constipation/diarrhea, relief from pneumonia, anti-inflammatory action on model mice with a type I allergy [1], anti-fatigue action on model mice during forced swimming [5].
However, efficacy mechanisms of OM-X® have not been fully examined. In this study, to examine OM-X® functions, in vitro studies using murine cells (gastrointestinal tract cells, RAW 264.7 cells) and in vivo studies using normal mice were conducted. The effects of orally administering OM-X® on functions of the liver gene expression, where absorbed OM-X® components are metabolized, were investigated.
Chemicals and Reagents: OM-X® (Lot.T101) was obtained from BIOBANK Co., Ltd., (Okayama, Japan). Lipopolysaccharide (LPS) was obtained from Macrophi Inc. (Shikoku, Japan). Dulbecco’s modified eagle’s medium and fetal bovine serum were purchased from Gibco Co., Ltd. (New York, USA). A rat GLP-1 cell kit was purchased from ReproCELL Co., Ltd. (Kanagawa, Japan). A GLP-1 active ELISA kit was purchased from Shibayagi Co., Ltd. (Gunma, Japan). A MILLIPLEX® MAP Mouse Cytokine/Chemokine Magnetic Bead Panel was purchased from Luminex Co., Ltd. (TX, USA). RNAlater® Solution was purchased from Thermo Fisher Scientific Inc. (MA, USA). An RNeasy Lipid Tissue Mini Kit was purchased from Qiagen Co., Ltd. (Venlo, Netherlands). An RNA 6000 Nano kit was purchased from Agilent Technologies Inc. (CA, USA). A MessageAmp biotin-enhanced amplification kit was purchased from Applied Biosystems Japan (Tokyo, Japan). All other laboratory chemicals were of the highest purity from commercial suppliers.
GLP-1 Production Assay in Lower Gastrointestinal Tract Cells: Lower gastrointestinal tract cells were isolated from Wistar rats and cultured in a 5% CO2 incubator at 37°C, as per the Rat GLP-1 cell kit instructions. After 12 h, OM-X® was added (final concentration 1–1,000 μg/ml, n = 3) and the supernatant was collected after culturing for an additional 40 min. The glucagon-like peptide-1 (active) (GLP-1) in the culture supernatant was quantified using the ELISA method.
NO and Cytokines Production Assay in RAW264.7 Cells: Macrophage (RAW264.7) cells suspended in a medium at 1.6 x 106 cells/ml were plated into the wells of a 96-well plate at 100 µl/well and cultured at 37°C in a CO2 incubator. Six hours later, OM-X® solution (final concentrations, 10–1,000 µg/ml; n = 2) or lipopolysaccharides (LPS) solution (final concentration, 8.0 ng/ml; n = 2) was added to the wells, and the culture was continued in the incubator. The culture supernatant was collected 24 h later, added to 100 µl Griess reagent (mixture of 7.5% phosphoric acid aqueous solution containing 3% sulfanilamide and 0.15% napthylethylenediamine solution in a ratio of 1:2), and incubated for 10 min, followed by measurement of absorbance by use of a microplate reader (primary wavelength, 550 nm; secondary wavelength, 750 nm). Similarly, cytokines in the obtained supernatant were determined using a MILLIPLEX® (TNF-α, IFN-γ, IL-1β, IL-4, IL-6, IL-12 (p40), IL-13, IL-17, MIP-1α) Panel Kit.
Oral Administration of OM-X® to Mice: OM-X® was dissolved in distilled water to give a concentration of 1% (w/v) solution, which was given ad libitum to the Six-week-old male ICR mice for 7 days (n = 5). In parallel, control mice were given distilled water only (n = 5). Finally, plasma and livers were harvested, and total liver RNA was extracted for focused DNA microarray analyses. All plasma was analyzed about Aspartate aminotransferase (AST) and Alanine Aminotransferase (ALT) levels. This study (protocol approval number: H27-010) conformed to the Guiding Principles for the Care and Use of Experimental Animals of Hokkaido Pharmaceutical University (published 1998, revised in 2001 and 2007).
RNA Extraction: Livers were dissected and stored in RNAlater® Solution for 7 days at −80°C. Total liver RNA was extracted (n = 2, OM-X® treatment group; n = 1, normal mice) using an RNeasy Lipid Tissue Mini Kit. The quality and quantity of the extracted RNA was assessed on a 2100 Bioanalyzer using an RNA 6000 Nano Kit according to the manufacturer’s instructions.
DNA Microarrays: cDNA was prepared from RNA and biotin-labeled RNA was transcribed and amplified using a MessageAmp Biotin-enhanced Amplification Kit according to the manufacturer’s instructions. Biotinylated amplified RNA (aRNA) was fragmented using fragmentation reagents and then incubated at 94°C for 7.5 min. The fragmentation reaction was terminated by the addition of the kit’s stop solution. The Innate Immunity Chip (183 genes), Anti-aging Chip (219 genes), and Metabolic Chip (195 genes) have on-surface chip probes and were used for the DNA microarrays. Hybridization signals were acquired using a DNA microarray reader with multi-beam excitation technology (Genopal®, Yokogawa Electric Co., Tokyo, Japan).
Statistical Analysis: GLP-1, NO, cytokines production, and blood marker results are expressed as the mean ± standard error of the mean (SEM). These tests were compared by performing one-way analysis of variance followed by Tukey’s test. For DNA microarray analysis, a 2.0-fold change in expression, as measured by the signal-intensity ratio (SIR = log2 [Sample Signal intensity/Control Signal intensity]), was interpreted as differentially-expressed genes (DEGs).
Pathway Analysis: The predicted target genes were compared against the Database for Annotation. Pathway analysis for DEGs was performed by referring to the latest Kyoto Encyclopedia of Genes and Genomes database (KEGG, http://www.genome.jp/kegg). This analysis enabled determining biological pathways significantly enriched in DEGs.
GLP-1 Production Assay in Lower Gastrointestinal Tract Cells: Significant increases in GLP-1 production were observed for rat lower gastrointestinal tract cells cultured in the presence of OM-X® at 1–1,000 µg/ml OM-X® relative to those for non-treated cells (Figure 1).
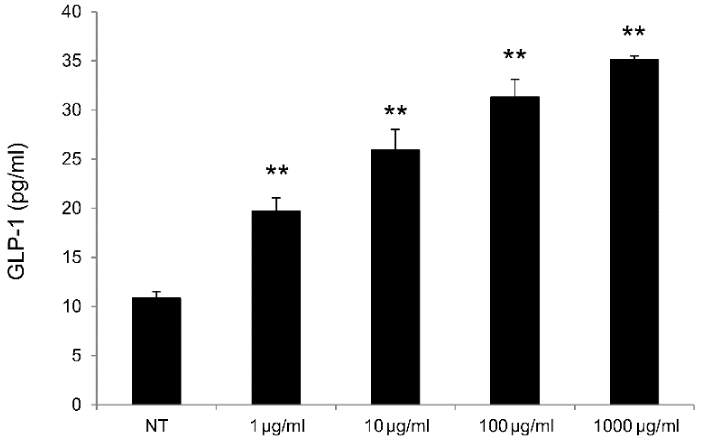
Figure 1. GLP-1 production by rat lower gastrointestinal tract cells treated with OM-X. OM-X was added to cultures at final concentrations of 1–1,000 (µg/ml), and the produced active GLP-1 in the supernatant was measured after 4 hours by ELISA. **P < 0.01, vs. NT (non-treatment).
NO and Cytokines Production Assay in RAW264.7 Cells: Addition of OM-X® to RAW264.7 cells led to a significant production of NO relative to that of the control (Medium; Med) at OM-X® concentrations of 100 and 1,000 µg/ml. Addition of OM-X® (1,000 µg/ml) plus LPS also resulted in significant production of NO relative to that of only LPS (Med + LPS) (Figure 2A).
Regarding cytokine production, addition of OM-X® (10–1,000 µg/ml) plus LPS led to significant production of IL-6 relative to that of addition of LPS (Med + LPS) (Figure 2B). Addition of OM-X® (100 µg/ml) resulted in significant production of TNF-α relative to that of the control (Med) and TNF-α level of OM-X® (1,000 µg/ml), and all additions of LPS were greater than the detection limit (10,000 pg/ml) (Figure 2C).
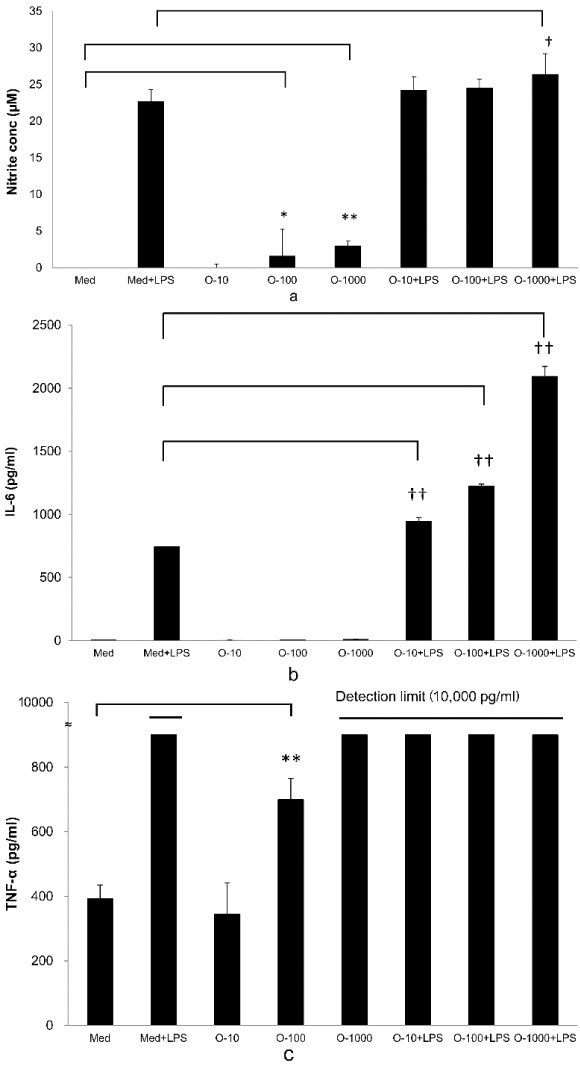
Figure 2. (A): NO production by RAW264.7 cells treated with OM-X for 24 hours. OM-X was added to cultures at final concentrations of 10–1,000 µg/ml or OM-X plus LPS (8 ng/ml). The produced NO in the supernatant was measured after 24 hours by Griess reagents (n = 2). *P < 0.05 ,**P < 0.01, vs. Med (Medium) , † P < 0.05, vs. Med + LPS. (B): IL-6 production by RAW264.7 cells treated with OM-X for 24 hours. OM-X was added to cultures at final concentrations of 10–1000 µg/ml or OM-X plus LPS (8 ng/ml). The produced IL-6 in the supernatant was measured after 24 hours using a MILLIPLEX panel kit (n = 2). †† P < 0.01, vs. Med (Medium) + LPS. (C): TNF-α production by RAW264.7 cells treated with OM-X for 24 hours. OM-X was added to cultures at final concentrations of 10–1,000 µg/ml or OM-X plus LPS (8 ng/ml). The produced TNF-α in the supernatant was measured after 24 hours using a MILLIPLEX panel (n = 2). TNF-α level of OM-X (1,000 µg/ml) and all additions of LPS were over the detection limit (10,000 pg/ml). **P < 0.01, vs. Med (Medium).
Plasma analysis and DNA Microarray Analysis of Gene Expression in the Liver: The AST and ALT of all groups were within normal levels (Data not shown).
The DNA microarray analysis of gene expression in OM-X®-administered mice livers identified the following 27 DEGs: 4 of 597 genes were upregulated, and 23 of 597 genes were downregulated (Figure 3) (Table 1).
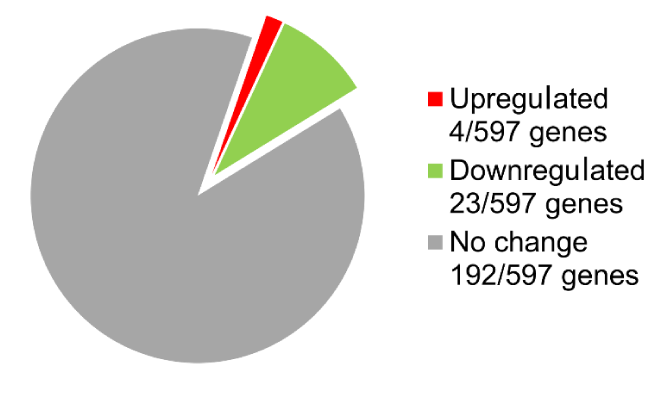
Figure 3. Analysis of gene expression changes in the liver of mice that orally received OM-X.
Number of DEGs (2.0-fold change) identified by the focused DNA microarray chip.
Table 1. List of 27 genes differentially expressed in OM-X treated mice determined using focused DNA microarray chips.
|
Gene symbol
|
Gene name
|
Signal intensity ratio
|
|
Traf1
|
TNF receptor-associated factor 1
|
10.2 ± 1.8
|
|
Traf2
|
TNF receptor-associated factor 2
|
9.4 ± 2.1
|
|
Socs4
|
suppressor of cytokine signaling 4
|
1.5 ± 0.1
|
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11
|
1.3 ± 0.1
|
|
Il1a
|
interleukin 1 alpha (Mus musculus)
|
−1.7 ± 0.7
|
|
Spp1
|
secreted phosphoprotein 1
|
−2.4 ± 0.5
|
|
Casp8
|
caspase 8
|
−2.4 ± 0.7
|
|
Il28a
|
interferon lambda 2
|
−3.7 ± 1.2
|
|
Ucp2
|
uncoupling protein 2
|
−6.8 ± 1.6
|
|
Map3k5
|
mitogen-activated protein kinase kinase 5
|
−8.1 ± 2.5
|
|
Tnf
|
tumor necrosis factor
|
−8.2 ± 1.2
|
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5
|
−9.5 ± 1.1
|
|
OmpA
|
outer membrane protein A
|
−9.7 ± 2.3
|
|
Casp4
|
caspase 4, apoptosis-related cysteine peptidase
|
−9.7 ± 2.4
|
|
Nlrp3
|
NLR family, pyrin domain containing 3
|
−9.8 ± 2.3
|
|
Ccl2
|
hemokine (C-C motif) ligand 2
|
−9.9 ± 2.3
|
|
Il28b
|
interferon lambda 3
|
−10.1 ± 0.6
|
|
Il1b
|
interleukin 1 beta
|
−10.4 ± 1.7
|
|
Ifng
|
interferon gamma
|
−10.5 ± 2.0
|
|
Zbp1
|
Z-DNA binding protein 1
|
−11.1 ± 1.5
|
|
Icam1
|
intercellular adhesion molecule 1
|
−11.1 ± 0.6
|
|
Bcl10
|
B cell leukemia/lymphoma 10
|
−11.3 ± 0.6
|
|
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10
|
−11.4 ± 1.5
|
|
Il12b
|
interleukin 12b
|
−11.5 ± 2.7
|
|
Il17ra
|
interleukin 17 receptor A
|
−12.2 ± 1.1
|
|
Casp9
|
caspase 9
|
−13.4 ± 0.6
|
|
Bcl6
|
B cell leukemia/lymphoma 6
|
−15.1 ± 0.6
|
The data is differential gene expression defined as a 2.0-fold change, which are shown as the signal intensity ratio (SIR) > 1.0 or < –1.0.
Pathway Analysis: Table 2 lists the diseases and pathways, which were obtained by means of the pathway analysis tools in KEGG Mapper, related to DEGs of OM-X®-administered mice livers.
Table 2. Pathway Search Result analyzed by KEGG software. DEGs of OM-X treated liver were found to be associated with inflammatory diseases and pathways.
|
Associated diseases or pathways
|
Associated gene number/DEGs
|
|
Influenza A
|
10/27
|
|
Cytokine-cytokine receptor interaction
|
9/27
|
|
Tuberculosis
|
8/27
|
|
TNF signaling pathway
|
8/27
|
|
Herpes simplex infection
|
8/27
|
|
Rheumatoid arthritis
|
7/27
|
|
Apoptosis
|
7/27
|
|
NF-kappa B signaling pathway
|
6/27
|
|
Non-alcoholic fatty liver disease (NAFLD)
|
6/27
|
|
AGE-RAGE signaling pathway in diabetic complications
|
5/27
|
|
Inflammatory bowel disease (IBD)
|
5/27
|
The results are computer predictions made by KEGG.
http://www.genome.jp/kegg-bin/color_pathway_object
2021 Copyright OAT. All rights reserv
To examine OM-X® functionality, the following points were considered. First, food components that are orally ingested are generally digested in the intestine, and some of the components are absorbed, metabolized in the liver, and enter blood circulation. Therefore, OM-X® may undergo similar digestion and absorption. Moreover, in the immune response to external substances, the innate immune system involving macrophages and dendritic cells is stimulated first; then, the immune signals are transmitted to T-lymphocytes and other cells to trigger a secondary immune response [6,7]. Thus, the effects of orally ingested OM-X® on the gastrointestinal tract, liver function, and innate immunity were evaluated in an exploratory manner.
Initially, the influence of OM-X® on GLP-1 productivity of rat lower gastrointestinal tract cells was examined. GLP-1 is produced in response to nutrient stimuli upon eating and contributes to blood glucose control by stimulating β cells in the pancreas to promote insulin secretion [8]. The study results showed that OM-X® increased GLP-1 production in a concentration-dependent manner. Because GLP-1 production is induced by nutrients, including glucose and lipids, OM-X® effects may be mediated by carbohydrates and lipids found in OM-X® [9].
Macrophage (RAW264.7 cells) has responsible for innate immunity. Recognition and elimination of foreign bodies by macrophages in infection and early stages of oncogenesis are critically important for preventing diseases. Toll-like receptors (TLRs) 2, 4, and 6 are involved in macrophage activation by pathogen-associated molecular patterns (PAMPs). For example, certain microbe-derived substances, including β-glucan and lipopolysaccharides, stimulate these receptors and thereby promote production of cytokines, like IL-1, IL-6, IL-12, and TNF-α [10,11].
In this study, the addition of OM-X® only or OM-X® plus LPS increased NO, IL-6, and TNF-α production. Accordingly, it is possible that OM-X® only or OM-X® plus LPS directly activate TLRs of macrophages and that this phenomenon is a priming action enhancing LPS stimulus by polysaccharide components in OM-X®. OM-X® contained 12 strains of LAB and bifidobacteria, including Lactobacillus acidophilus, L. brevis, L. bulgaricus, L. casei, L. fermentum, L. helveticus, L. plantarum, Bifidobacterium breve, B. longum, B. infantis, Streptococcus thermophilus, and Enterococcus faecalis TH10. Especially, E. faecalis TH10 of OM-X® is an LAB strain isolated from tempeh, a traditional fermented food in Southeast Asia. Therefore, PAMPs of bacteria from OM-X® stimulate TLRs and induce many kinds of cytokines [12].
Finally, a focused DNA microarray (Genopal®) was used to analyze changes in genes expressed in the liver as a hepatic effect of orally administered OM-X® in mice [13].
The information of liver genes that were differentially expressed in response to OM-X® administration was analyzed using pathway analysis tools in KEGG Mapper. Thus, it was predicted depending on strong suppression of genes, except Traf1/2, that OM-X® is involved in infectious diseases (influenza A, tuberculosis, Herpes simplex infection), inflammatory diseases (rheumatoid arthritis, NAFLED, diabetic complications, IBD), and inflammation-related pathways (TNF signaling, NF-kappa B) and acts on these inflammatory reactions in a suppressive manner [14,15,16]. Given the results indicating that OM-X® tends to suppress apoptosis pathways, OM-X® can be expected to have a cytoprotective action on hepatocytes through suppression of their cell death, which occurs in response to various infections and inflammatory reactions [17,18].
Moreover, OM-X® orally administered to mice suppresses inflammatory genes, including TNF in the liver, while having a direct stimulatory action on macrophages to enhance their TNF-α production, which indicates that OM-X® is a biological response modifier with an immunomodulatory activity [19,20,21,22].
We appreciate the assistance of Mr. Naoyuki Togawa (Bio-device Research group, Mitsubishi Rayon. Co., Ltd., Tokyo, Japan) who provided technical information on DNA microarray analysis during the study period. This study was funded by BIOBANK Co., Ltd. (Okayama, Japan).
None
- Itoh T, Miyake Y, Kasashima T, Shimomiya Y, Nakamura Y, Ando M, et al. (2015) OM-X®, Fermented Vegetables Extract Suppresses Antigen-Stimulated Degranulation in Rat Basophilic Leukemia RBL-2H3 Cells and Passive Cutaneous Anaphylaxis Reaction in Mice. Nat Prod Commun 10: 1597-1601.
- Spierings EL, Walshe T, Pescatore F (2013) Safety and tolerability of Dr Ohhira’s OM-X capsules in Healthy volunteers. Integr Med 12: 38-42.
- Takahata M, Frémont M, Desreumaux P, Rousseaux C, Dubuquoy C, Shimomiya Y, et al. (2014) Evaluation of therapeutic properties of fermented vegetables extract (OM-X) in the model of colitis induced by Citrobacter rodentium in mice. J Funct Foods10: 117-27.
- Kawakami M, Ohhira I, Araki N, Inokihara K, Matsubara T, Iwasaki H (1998) The influences on the VO2max of athlete with taking lactic acid bacteria (OM-X). Kurashiki University of Science and the Arts 3: 131-44.
- Oszmianski J, Lachowicz S (2016) Effect of the Production of Dried Fruits and Juice from Chokeberry (Aronia melanocarpa L.) on the Content and Antioxidative Activity of Bioactive Compounds. Molecules 21: 1098.
- Wu G, Johnson SK, Bornman JF, Bennett SJ, Fang Z (2017) Changes in whole grain polyphenols and antioxidant activity of six sorghum genotypes under different irrigation treatments. Food Chem 214: 199-207.
- Abuajah CI, Ogbonna AC, Osuji CM (2015) Functional components and medicinal properties of food: a review. J Food Sci Technol 52: 2522-2529. [Crossref]
- Berraondo P, Minute L, Ajona D, et al. (2016) Innate immune mediators in cancer: between defense and resistance. Immunol Rev 274: 290-306. [Crossref]
- Muszynski JA, Thakkar R, Hall MW (2016) Inflammation and innate immune function in critical illness. Curr Opin Pediatr 28: 267-273. [Crossref]
- Said H, Kaunitz JD (2016) Gastrointestinal defense mechanisms. Curr Opin Gastroenterol 32: 461-466. [Crossref]
- Arango Duque G, Descoteaux A (2014) Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5: 491. [Crossref]
- Borges PV, Moret KH, Raghavendra NM, Maramaldo Costa TE, Monteiro AP, Carneiro AB, et al. (2016) Protective effect of gedunin on TLR-mediated inflammation by modulation of inflammasome activation and cytokine production: Evidence of a multitarget compound. Pharmacol Res 115: 65-77.
- Itoh T, Miyake Y, Onda A, Kubo J, Ando M, Tsukamasa Y, et al. Immunomodulatory effects of heat-killed Enterococcus faecalis TH10 on murine macrophage cells. Microbiologyopen 1: 373-380.
- Taira M, Nezu T, Sasaki M, Kimura S, Kagiya T, Harada H, et al. (2009) Gene expression analyses of human macrophage phagocytizing sub-micro titanium particles by allergy DNA chip (Genopal). Biomed Mater Eng 19: 63-70.
- Pinar A, Dowling JK, Bitto NJ, Robertson AB, Latz E, et al. (2016) PB1-F2 derived from avian influenza a virus H7N9 induces inflammation via activation of the NLRP3 inflammasome. J Biol Chem [Crossref]
- Marques CD, Dantas AT, Fragoso TS, Duarte AL (2010) The importance of vitamin D levels in autoimmune diseases. Rev Bras Reumatol 50: 67-80. [Crossref]
- Lee JH, Lee B, Lee HS, Bae EA, Lee H, Ahn YT, et al. (2009) Lactobacillus suntoryeus inhibits pro-inflammatory cytokine expression and TLR-4-linked NF-kappaB activation in experimental colitis. Int J Colorectal Dis 24: 231-237.
- Choi MR, Do le T, Chung YH, Yoo H, Yu R (2015) Antioxidative Activity of Platinum Nanocolloid and Its Protective Effect Against Chemical-Induced Hepatic Cellular Damage. J Nanosci Nanotechnol 15: 5571-5576. [Crossref]
- Ma JQ, Ding J, Zhang L, Liu CM (2014) Hepatoprotective properties of sesamin against CCl4 induced oxidative stress-mediated apoptosis in mice via JNK pathway. Food Chem Toxicol 64: 41-48. [Crossref]
- Huang LH, Pan XP, Gong KR, Shao G. Anti-inflammatory effects of three kinds of traditional Mongolian medicine monomer and its combination on LPS-stimulated RAW264.7 macrophages. Eur Rev Med Pharmacol Sci 20: 950-958.
- Yang G, Ham I, Choi HY (2013) Anti-inflammatory effect of prunetin via the suppression of NF-κB pathway. Food Chem Toxicol 58: 124-132. [Crossref]
- Zheng X, Zou S, Xu H, Liu Q, Song J, et al. (2016) The linear structure of I²-glucan from baker's yeast and its activation of macrophage-like RAW264.7 cells. Carbohydr Polym 148: 61-68. [Crossref]