Abstract
Using CRISPR/Cas9 genome-editing technique, human blood cell-derived iPS cell clones lacking HLA class I expression were established. Considering the risk for the “off-target” effect, D10A nickase type mutant Cas9, as well as wild type (WT) Cas9, were utilized with sgRNA designed for sense and anti-sense strand of β2-microglobulin (β2m) gene loci. One pair of designed sgRNA provided highly efficient genome editing, consistent with the observation that the sgRNA efficiently reconstituted EGFP expression by homology dependent repair of pCAG-EGxxFP reporter plasmid. Obtained HLA class I-lacking iPS cells showed intact pluripotency and off-target mutations seemed to be very rare. These HLA class I-deficient iPS cells may be useful to study the host immune reactions by some innate immune cells, as well as T cells for the future clinical applications of stocked iPS cells.
Key words
β-2 microglobulin, CRISPR/Cas9, HLA class I, iPS cell
Introduction
Induced pluripotent stem (iPS) cells [1] and genome editing technique using clustered regulatory interspaced short palindromic repeat (CRISPR)-associated (Cas) system [2-5] are the most epoch-making advancement in the field of life science in recent decades. iPS cells have great potential for basic disease research including drug estimation and clinical application as regenerative medicine. Patient-derived iPS cells, which are induced to differentiate into various tissues or cells, are expected to escape from host immune reactions when they are transplanted into the each patient. However, establishment and safety considerations of the personalized iPS cells require vast time and cost. Therefore, it has been proposed that iPS cell bank should be constructed [6].
Human Leukocyte Antigens (HLA), encoded by the genes located in the major histocompatibility complex on chromosome 6, are the major target molecules for the host immune system when the tissues or the cells are transplanted. Among them, HLA class I is expressed on almost all tissue cell types and consists of highly polymorphic heavy chain (including HLA-A, HLA-B and HLA-C) and invariant light chain, 1. Polymorphic HLA class I antigens are distinguished by mainly host CD8+ T lymphocytes and transplants are attacked by them, resulting in the rejection. While HLA type should be identical between donor and host to avoid the rejection, complete matched subject can not be always obtained because of quite high polymorphism of the HLA gene.
Since invariant light chain β2m is essential for the surface expression of HLA class I antigens, destruction of β2m gene results in almost complete loss of HLA class I expression [7]. Therefore, β2m-null iPS cells and differentiated cells from them might reduce the immunogenicity and consequently escape from the attack of host T cells. In this study, we established β2m-null iPS clones using a recent powerful genome editing technology, CRISPR/Cas9 system. Considering the risk for “off-target” effect, in which Cas9 nuclease tolerates certain mismatches resulting in undesired mutagenesis, we utilized nickase type mutant Cas9, Cas9n(D10A), as well as wild type (WT) Cas9 [8,9]. Established novel β2m-null iPS cells may provide useful tools to examine whether lacking of HLA class I expression of iPS-derived transplants promote successful engraftment.
Materials and methods
Human iPS cells generation
Mononuclear cells were obtained from peripheral blood of healthy volunteer using BD vacutainer (BD 362760). Collected cells were stimulated by anti-CD3/CD28 coated beads (Velitas DB11131) in the presence of rIL-2 (Novartis). The activated cells were transduced with reprogramming factors (Oct4, Sox2, Klf4 and c-myc) via retrovirus vector, which were kindly provided by Drs. Hiromitsu Nakauchi and Shin Kaneko (The Institute of Medical Science, Tokyo University, Tokyo, Japan). Infected cells were plated on mitomycin C (MMC)-treated mouse embryonic fibroblasts (MEF) in RPMI 1640 medium containing 10% FBS, which was gradually replaced with iPS medium, 80% DMEM F12 (Sigma D6421), 20% Knock Out serum (GIBCO 10828-028 ), 1% Non essential amino acids (GIBCO 11140), 1x L-Glutamate Penicillin Streptmycin (sigma G1146 ) and 5ng/ml of bFGF (Wako 060-04543). The established iPS colonies were picked up and expanded.
Cell culture
iPS cells were cultured on MMC-treated MEF in iPS medium and passaged every 3 or 4 days in the presence of 10 ug/ml of Y27632 (Wako 251-00514). HEK293 cells were cultured in DMEM (Nissui 05918) containing 10% FBS and 1x L-Glutamate Penicillin Streptmycin (sigma G1146 ).
sgRNA and Cas9 mRNA preparation
2 guide RNAs targeting human β2m gene exon 1 and 2 were selected by optimized CRISPR DESIGN (http://crispr.mit.edu). sgRNAs were designed by ligation of tracrRNA sequence to 3’-end of the guide RNA sequences. Double strand DNA fragments containing T7 promoter and sgRNAs were synthesized. Cas9 WT or Cas9n(D10A) with or without IRES-mCherry sequence was cloned into R522 vector, which harbors CMV promoter/enhancer, rabbit beta-globin intron and polyA signal sequence. mCherry coding sequence was derived from pmCherry-N1 vector, purchased from Clontech (632523). Using mMESSAGE mMACHINE T7 ULTRA kit (ambion AM1345), sgRNA and Cas9 mRNA were in vitro transcribed. And then they were purified by MEGA clear kit (ambion AM1908). To examine whether sgRNA has functional activity in vitro, PCR-amplified genomic fragments of β2m gene exon 1 or 2 were incubated with synthesized sgRNA and recombinant Cas9 (NEB M386) at 37℃ for 1 hour and electrophoresed to confirm the digested fragments.
Validation of sgRNAs and Cas9 mRNA in vivo
pCAG-EGxxFP, pCAG-EGxxFP-Cetn1, and pX330-Cetn1/1 were given from Dr. Ikawa through Addgene (Plasmid ID #50716, #50717 and #50718, respectively.) (10, 11) Genomic fragments containing sgRNA target sites fo β2m exon1 (547 bp) and exon 2 (535 bp) were amplified by PCR and were inserted into pCAG-EGxxFP EcoRI/SalI site, which were co-transfected to HEK293 cells using lipofectamin (Invitrogen 18324-012) with in vitro transcribed sgRNAs and Cas9 WT-IRES-mCherry or Cas9n(D10A)-IRES-mCherry mRNA. 48 hours after transfection, reconstituted EGFP expression by homology dependent repair within mCherry-positive cells was analyzed by a flowcytometer (BD LSRFoetessa).
Establishment of β2m-null iPS clones
Before CRISPR/Cas9 treatment, iPS cells cultured on MEF were transferred to feeder-less culture condition. Briefly, single cell suspension of iPS cells were re-suspended into ReproXF (Reprocell RCHEM007) medium containing bFGF(10 ug/ml) and ROCK inhibitor (10 uM) then plated onto geltrex (GIBCO A1413301) coated well. After several days feeder-less culture, cells were collected using accutase (Thermo Fisher A1110501) and 2.8 x 10 [5] cells were re-suspended into 10 ul of Opti-MEM medium (GIBCO 31985-070 ). Then cells were electroporated with Cas9 mRNA and sgRNAs using Neon transfection system (Thermo Fisher) under the condition that pulse voltage is 1000 V, pulse width is 30 ms and pulse number is 1. After electroporation, cells were cultured on MEF with iPS medium. After the culture, collected cells were stained with FITC-labeled anti-HLA-ABC (clone G46-2-6, BD Pharmingen 555552) and negative cells were sorted by FACSAria (BD). Sorted or non-sorted cells were cultured on MEF with iPS medium and colonies were isolated. β2m gene exon 1 region was amplified from genome DNA of each clone and nucleotide sequence was analyzed by Sanger method using a genetic analyzer (Thermo Fisher 3500XL).
Examination of off-target effect
Candidate off-target sites were searched using optimized CRISPR DESIGN (http://crispr.mit.edu). Among candidate 164 off-target sites, 11 sites which are located within protein coding region (CDR) were amplified by PCR using genome DNA of established iPS null clones (Cas9 WT-mediated S1-3 and Cas9n(D10A)-mediated S3-7) as templates and nucleotide sequences were analyzed by Sanger method using a genetic analyzer (Thermo Fisher 3500XL).
Teratoma formation
To examine pluripotency in vivo, single suspension of 5x10 [5] of β2m -/- iPS cells in 5 ul DMEM were transplanted between the kidney capsule and the renal cortex using fine tip glass pipette. 6 weeks later, recipient mice were given an overdose of pentobarbital (60 mg/kg, i.p.), and were perfused with warm 0.01M phosphate-buffered saline (PBS) through the left ventricle, followed by fixation with 4% paraformaldehyde/0.1M phosphate buffer (4% PFA/PB). Kidneys were then removed and re-fixed overnight in4%PFA/PB, washed with graded sucrose 0–25%/0.01M PBS series, and were quick frozen in isopentane pre-cooled with liquid nitrogen, followed by storage at −80°C until use. For the histological staining, several 7-µm sections were obtained. Engrafted human cells were detected by anti-human nuclear antigen (HNA, 1:100, 4°C overnight; clone 235-1, Cy3 conjugate, Millipore, Temecula, CA). Cell differentiation into endoderm, ectoderm and mesoderm were confirmed using biotin conjugated rabbit polyclonal anti-E-Cadherin (1:50, 4°C overnight, eBioscience 13-3249 clone: DECMA-1, San Diego, CA), as a detection of epidermal cell, rabbit polyclonal anti-GFAP (1:100, 4°C overnight, LAB VISON, RB-087-A, Fremont, CA), as of glial cells, and rabbit polyclonal anti-skeletal muscle actin (1:100, 1 h at room temperature, Abcam, ab15263, Cambridge, UK) as of skeletal muscle cell, respectively. Reactions were visualized using Alexa Fluor-488-conjugated goat anti-rabbit and anti-rat antibodies (1:500, for 2 h at room temperature; Molecular Probes, Eugene, OR). Nuclei were counter-stained with DAPI (4’,6-diamino-2-phenylindole, VECTOR LABORATORY H-1200). All animal experiments were approved by the Tokai University School of Medicine Committee on Animal Care and Use (Kanagawa, Japan), and were performed in the Tokai University School of Medicine.
Results
Design and validation of sgRNAs with Cas9 WT and Cas9n(D10A) mutant
Cas9 nuclease, derived from Streptococcus pyogenes, forms the complex with a sgRNA that hybridizes a ~20 nucleotides DNA immediately preceding the “PAM” motif (NGG) and makes double strand break (DSB) at 3 bp upstream of the PAM site. This system, called “CRISPR-Cas9 system”, has been widely used in lots of recent reports to edit desired genomic sites with high efficiency in mammalian zygotes and cultured cells system [2-5]. However, it has been concerned that the system affects on undesired genome sites at some frequency (off-target effect). On the other hand, Cas9 D10A mutant, Cas9n(D10A), makes a nick on a single strand DNA but not DSB. Using two sgRNAs designed for sense and anti-sense strands within a several base pairs, Cas9n(D10A) cuts each single strand and consequently forms DSB with some cohesive sequences. It is suggested that the rate of off-target effect is reduced when Cas9n(D10A) system is used compared to Cas9 WT [8,9].
We designed two guide RNA pairs for β2m gene exon 1 and 2 regions (Figure 1). Then we synthesized DNA containing the sequences for T7 promoter, designed guide RNA described above and tracrRNA which is involved in the complex formation with Cas9. Using these DNA fragments as templates, “sgRNA” can be transcribed in vitro by T7 RNA polymerase. We initially confirmed that DNA fragments containing target sequences either at exon 1 or 2 were evidently digested with in vitro transcribed sgRNA and recombinant Cas9 in vitro (data not shown). Next, we validated sgRNA in vivo using an EGFP expression cassette which is to be reconstituted with DSB at an inserted target sequence and homology dependent repair [10,11]. Hela cells were transfected with the EGFP expression cassette containing β2-microglobulin gene exon 1 or exon 2 region and DNA vectors expressing designed sgRNA and Cas9-IRES-mCherry. As shown in Figure 2, 12-23% of the cells expressed mCherry and significant fraction of the mCherry-positive cells expressed EGFP when sgRNA for exon 1 was transfected but not when sgRNA for exon 2 was transfected. Cas9 WT and D10A mutant showed similar result.
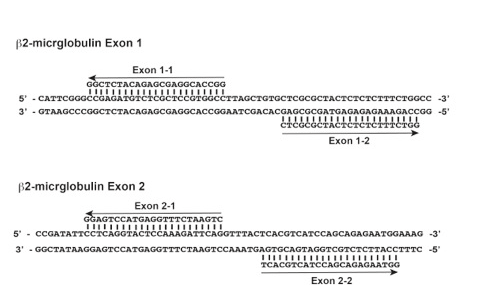
Figure 1. Guide RNA sequences for human b2-microglobulin exon 1 and 2 locus. Each guide RNA pairs are separated by 9 and 6 bp, respectively. PAM sequences (NGG) of the guide RNA pair are out-oriented.
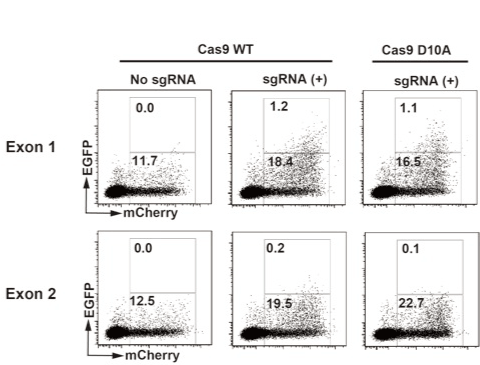
Figure 2. Validation of sgRNAs using EGxxFP system.
sgRNAs for exon1/2 and mRNAs for Cas9 IRES mCherry were in vitro transcribed. Synthesized RNAs and pCAG EGxxFP b2m exon1 or pCAG EGxxFP b2m exon2 were transfected to HEK293 cells. After 48 hours, cells were analyzed by a flowcytometer. Data were analyzed using Flow Jo 7.6 (Tomy Digital Biology). Values are percentages of boxed regions.
Production of β2-microglobulin gene-modified human iPS cells
We have established iPS cells from peripheral blood T cells of healthy volunteer. One of the clones, designated clone 441, was prepared as single cell suspension and transfected with Cas9 WT or Cas9n (D10A) mRNA along with sgRNA pair for exon 1 or 2 using Neon electroporation system. After the electroporation procedure, cells were cultured on mouse embryonic fibroblast (MEF) with conventional iPS cell culture media. Several days later, cells were collected and applied to a flowcytometry analysis. Exon 1 sgRNA-treated iPS cells showed decreased HLA class I (HLA-ABC) expression whereas exon 2 sgRNA-treated cells did not (Figure 3), suggesting that the destruction of β2m gene through exon 1 sgRNA plus Cas9 results in the loss of surface expression of HLA-class I. Decreased expression of HLA class I was observed in both iPS cells treated with Cas9 WT and Cas9n (D10A). This result was consistent with the EGFP expression experiment, showing that the sgRNA for exon 1 was more effective for genome editing than for exon 2 and that the frequency was not different between Cas9 WT-treated and Cas9n(D10A)-treated cells (Figure 2). Then, clones were isolated before or after the sorting of HLA class I-negative fractions. All clones derived from exon 1-treated cells were HLA class I-negative (Figure 4, and data not shown). Next, we confirmed the genome sequence of the obtained clones at β2m exon 1 region (Table 1). All clones tested harbored mutated alleles on both chromosomes, indicating that genome editing by CRISPR/Cas9 system works quite efficiently when appropriate sgRNA was selected.
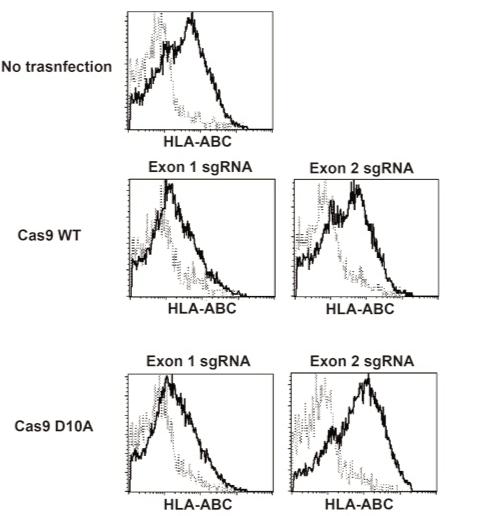
Figure 3. Wild type and D10A nickase Cas9 mutant efficiently generated HLA class I-deficient iPS cells.
sgRNAs and Cas9 WT or Cas9n(D10A) transfected iPS cells were stained with FITC-labeled anti HLA-ABC antibody (clone G46-2-6). Data were analyzed using CellQuest Pro program (BD Bioscience).
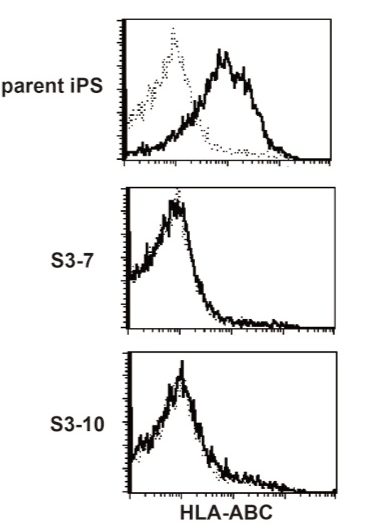
Figure 4. HLA class I-deficient iPS clones.
Cloned CRISPR treated iPS cells were stained with the anti human HLA ABC antibody. Typical result was shown, but all other clones tested were HLA ABC-negative.
| |
|
|
|
|
|
b2m Ex.1 original sequence
|
|
|
|
clone
|
CGGGCCGAGATGTCTCGCTCCGTGGCCTTAGCTGTGCTCGCGCTACTCTCTCTTTCTGGCCTGGAG
|
|
|
|
S3-10
|
GTGGC---------------acagCTCTC
|
15bp(-) 4bp(+)
|
frame shift
|
|
|
GCTACcgccttagctgtgctcgcgctacTCTCT
|
23bp(+)
|
frame shift
|
|
S3-7
|
--------------
|
14bp(-)
|
frame shift
|
|
|
------------------------
|
24bp(-)
|
|
|
P3-5
|
GCGCTccgtggccttagctgtgctcgcgctACTCT
|
25bp(+)
|
frame shift
|
|
|
-----
|
5bp(-)
|
frame shift
|
|
S1-2
|
---------------
|
15bp(-)
|
start codon missing
|
|
|
---------------
|
15bp(-)
|
start codon missing
|
|
S3-4
|
-----------
|
11bp(-)
|
frame shift
|
|
|
---------
|
9bp(-)
|
|
|
S3-6
|
----------
|
10bp(-)
|
frame shift
|
|
|
CTCCGaGGCCTTAtCTGTGgTCGggctactctgtgCGCTAC
|
12bp(+), 3MM
|
|
|
S3-7
|
TGGCCgcacccgcgccctcctgtctctctccgtggccttagctgtgCTCGC
|
41bp(+)
|
stop codon
|
|
|
--------------
|
14bp(-)
|
frame shift
|
Table 1. Nucleotide sequence of b2m exon 1 region in genome-edited iPS clones. Underlined sequences are target sites for sgRNAs. "-" correponds to deleted nucleotides. Lower case characters are inserted nucleotides, whereas upper case characters are nucleotides identical to the original sequence. Red letters corespond to start and stop codons.
Quality estimation of genome-edited iPS clones.
In order to confirm that the genome editing procedure did not impair the pluripotency of iPS cells, β2m null iPS clone (clone S3-7) was transplanted under the kidney capsule of athymic nude mouse. After 3 months, β2m null iPS cells formed teratoma (Figure 5), in which all three embryonic layers were developed. E-cadherin expressing epithelial-like cells (endoderm), GFAP-expressing neural cells (ectoderm) and skeletal muscle actin (SK-actin)-expressing myocyte lineage cells (mesoderm) were detected, suggesting that the genome edited iPS cell retains pluripotency.
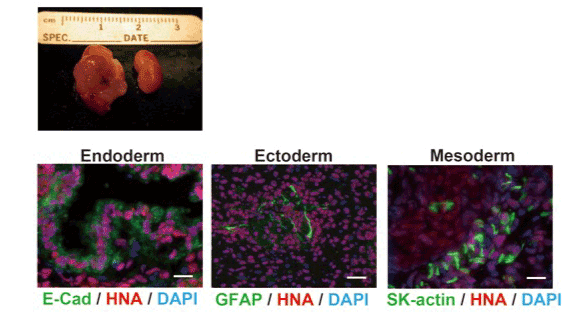
Figure 5. Established HLA class I-deficient iPS cells maintained pluripotency.
To investigate whether established HLA class I-deficient iPS cells retain pluripotency, t iPS cells were transplanted into nude mouse kidney capsule. After 6 weeks, mice had been sacrificed. Tumor had developed only in transplanted kidney (left) but not in un-treated kidney (right). Tissue sections were stained with anti E-Cadherin antibody (E-Cad) as a marker of endoderm, anti GFAP antibody as a marker of ectoderm, and anti Skeletal Muscle Actin antibody (SK-actin) as a marker of mesoderm. All sections were stained with anti human nuclei antibody (HNA) to show that the cells were human origin. Scale bars are 20 (left), 50 (middle) and 20 (right) µm.
Next, we examined whether “off-target” mutations arose in the β2m null iPS cells. 164 candidate off-target sites for Ex1-1 and Ex1-2 sgRNAs are computationally selected. 15 loci are 3 base mismatched and 149 are 4 base mismatched. Among them, off-target sites which are located within protein coding region (CDR) are 4 (3 mismatched) and 7 (4 mismatched) (Table 2). Nucleotide sequences of these 11 candidate off-target sites are examined in clones S1-2 (Cas9 WT-mediated) and S3-7 (Cas9n (D10A)-mediated). We found no alterations in both clones, suggesting that off-target mutations attributable to Cas9 WT and D10A are very rare.
In this study, we established HLA class I-lacking iPS clones using CRISPR/Cas9 technology, which was quite efficient when appropriate sgRNA was selected and the frequency of undesired off-target mutation was relatively lower than expected. Selection of sgRNA seems quite important. Although sgRNA for β2m exon 2 fully digested target sequence with recombinant Cas9 in vitro, these sgRNA did not function in living cells for unknown reason. On the contrary, EGFP reconstitution assay using the EGxxFP reporter plasmid in in vivo condition indicated that sgRNA for β2m exon 1 was more effective than that for exon 2. For the appropriate selection of sgRNA, EGFP reconstitution assay seems to be useful,
Emerging genome editing technique through CRISPR/Cas9 system may provide increased chance of iPS cells to study and to cure several disorders. Comparison of some disease patient-derived iPS cells with those derived from healthy volunteer may clarify some causative phenotypes for the disease. However, the possibility that the phenotype is resulted only from different genetic background or varied character of iPS clones, but not from disease itself, should be excluded. For such purpose, CRISPR/Cas9 is a powerful tool, enabling us to directly compare the relevant gene-edited or non-edited iPS clones, either of patient-derived or healthy control-derived.
HLA class I plays predominant role in transplantation rejection, in which CD8 T cells are mainly involved. Therefore, the attack by CD8 T cells could be avoided if HLA class I expression is genetically disrupted. On the other hand, it is well known that some HLA class I molecules deliver an inhibitory signal into natural killer (NK) cells and consequently that tumor cells lacking HLA class I are attacked by natural killer (NK) cells. Whether genetically engineering of HLA gene in iPS cells could contribute to augmentation of regenerative medicine is to be further investigated.
We thank Dr. Shin Kaneko (CiRA, Kyoto University) for instruction of iPS cell generation; Mr. Yoshinori Okada and Ms. Yumi Iida (Support Center for Medical Research and Education, Tokai University) for assistance of flowcytometer operation; Mr. Hideki Hayashi and Mr. Masayuki Tanaka (Support Center for Medical Research and Education, Tokai University) for assistance of computational analysis; Ms. Maki Masuda (Division of Basic Medical Science and Molecular Medicine, Tokai University) for assistance of histological experiment.
The authors declare that they have no competing interests.





