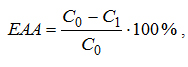Antagonistic qualities of lactic acid bacteria (LAB), including Bifidobacteria are well known. This primarily concerns a wide variety of pathogenic microorganisms that cause both food spoilage and human diseases of varying severity. Such qualities are explained by LAB production of substances called bacteriocins. There is a certain relationship between cancer and microbes, bacteria and viruses. However, information on the possibility of cancer cell inhibition by means of their effect on activating microorganisms is practically absent. The present study suggests four active strains which have been isolated from the gastrointestinal tract of human New isolates SK1, SK2, SK8 and SK11 have been identified as Bifidobacterium. A wide range of antioxidant, antimicrobial and anticancer properties against the four strains of Bifidobacterium SK and cellular test cultures was studied. It was found that the strains of Bifidobacterium SK1, SK2, SK8, SK11 completely inhibit the growth of E. coli В-6954, Staphylococcus aureus ATCC 25923, Salmonella enterica ATCC 14028, Klebsiella pneumoniae В-7001. In relation to Clostridium tyrobutyricum LMG inhibitory activity was observed only in SK2 strain. The antitumor activity of Bifidobacterium isolates determines the promising direction of their use in the prevention and in the immediate treatment of cancer. For example, as the basis for product recovery of cancer patients in remission after chemotherapy.
Anti-cancer, LAB, Bioactive microbial metabolites, Bacteriocins
One of the main causes of peoples’ death, regardless of economic development, is cancer [1,2]. Existing medical technology enables to achieve a complete recovery only in limited cases, and usually in the presence of early diagnosis. A significant role in the recovery is given to the rehabilitation technologies during or after chemotherapy, to the side effects that is a serious problem [3,4]. It is vital to develop technologies that allow both to carry out prevention and to reduce risk of death and disability of cancer patients.
There are papers in which the relationship between cancer and microbes, bacteria and viruses [5-11] are studied, but information about the possibility of inhibition of cancer cells by means of exposure to microbes activates them are practically absent. As potential effects of agents can be considered lactic acid bacteria that have shown inhibitory activity against pathogenic bacteria, fungi and yeast in vitro studies [12-24].
The main species of lactic acid bacteria (LAB) are Lactobacillus and Bifidobacterium, Enterococcus and Saccharomyces [25-30]. There is a centuries-old tradition of their use in the food, agriculture and processing industry, medicine, biotechnology, cosmetics, etc. [31-39]. Lactobacteria are a group of organisms most frequently used as the activity of lactic acid bacteria determines the possibility of their use as a cultural preserve [40]. A wide range of antimicrobial, antifungal activity of lactic acid bacteria determines the possibility of their use in bio-conservation technologies [15,20-23,41-48].
In order to solve practical problems a large initial screening test of different cultures requires to find active lactobacillus responsible specified quality.
In the present study four active strains, previously isolated from the gastrointestinal tract of human were studied. The aim was to identify and characterize their antioxidant and antimicrobial properties, determined by allocated bacteriocins and set antitumor activity of these metabolites. The importance of taxonomic characterization and identification of the isolates is defined by insufficient knowledge of the spectrum of the human microbiota and mechanisms of action of bacteriocins on cell and microbial culture. We have studied in vitro potential of isolated Bifidobacterium and their bacteriocins to inhibit cancer of cell cultures
Lactic acid bacteria isolates, media and cultivation conditions
4 strains of lactic acid bacteria, identified as SK1, SK2, SK8, SK11 were pre-selected for this study. They have been allocated from the gastrointestinal tract of 500 healthy and cancer patients in different age groups (Russian Federation). Pure cultures were stored at 4 ± 2°C in a lyophilized form. Before determination of the strains they were pre-cultured twice under anaerobic conditions (Biostat A plus MO fermenter, Sartorius, USA) in MRS-broth for 24 hours at 37°C.
Overnight culture of micro-organisms were collected by centrifugation (3500 rev/min, 30 min, 4°C) and washed twice with phosphate saline buffered (PBS). The cell concentration was adjusted to 105 CFU/ml, respectively, and the cells were obtained by heating the bacteria to 95°C for 1 hour. After the heat treatment, the cells were washed with PBS and resuspended in a suitable medium for the analysis of their viability. For fractionating of cell culture was centrifuged (3500 rev/min, 30 min, 4°С). The cell pellet was washed and resuspended in 100 ml PBS for 20 min (minute intervals), sonication was performed in a chilled water (4°С). Cell debris was removed by centrifugation (14 000 rev/min, 1 h), and supernatants were sterilized by filtration (pore size is 0.22 nm; Sartorious, Goettingen, Germany).
Exponential LAB cultures in MRS broth were used as inoculum for the antimicrobial, antioxidant and anti-cancer tests as follows: cancer and test cultures cells were grown with cells suspensions containing the collected cells of lactic acid bacteria (~105 CFU/ml of log phase growth of the culture of each strain) for 24 hours and then transferred into the chamber.
The test cultures, media and growing conditions
test strains were used: E. coli В-6954, Staphylococcus aureus ATCC 25923, Salmonella enterica ATCC 14028, Klebsiella pneumoniae В-7001, Listeria innocua LMG, Clostridium tyrobutyricum LMG, Lactobacillus plantarum, Lactobacillus acidophilus, Lactobacillus rhamnosus, Lactobacillus paracasei, Lactobacillus fermentum, Lactobacillus salivarius, Lactobacillus casei, Lactobacillus reuteri, Streptococcus agalactiae, Enterococcus faecium, from the laboratory collection of Research Institute Biotechnology of Kemerovo Institute of Food Science and Technology (University). The slurry of night broth cultures of the test strains grown on standard nutrient media was taken for the work. The number of microorganisms (titer) in the suspension was determined by optical density (OD) at a wavelength of 595 nm.
Test cultures were grown on meat-peptone agar (MPA) for 24 hours at 37°С; Cells from the agar surface were collected with microbiological loop and resuspended in NaCl solution up to 109. Bacilli were grown in MRS-broth for 24 hours at 37°C, then the culture liquid was centrifuged at 2000 rev/min for 10 min and the supernatant was separated. For the cell separation, supernatant was filtered through Millex-GV (0.22 um, Nihon «Millipore», USA). The plate was added with 180 ul of each culture of Bacillus genus, then 20 ul of each test strain was added to the cultures. The plate was incubated at 37°C for 24 h. As controls, 200 ul of sterile MRS-broth and 180 ul of pure medium with 20 ul of a solution of each pathogen were used. Bacterial growth was monitored by measuring the optical density during culturing [31].
To examine antagonistic activity of lactic acid bacteria, MRS medium and Lysogeny broth (LB) were used. The components of culture media were dissolved in 1 dm³ of the distilled water, heated until complete melting of the agar, distributed into tubes or flasks and sterilized for 1 hour at 121°C.
Cell culture and adhesion of cancer cells
As test cultures the following cancer cells from the collection of Cancer Center Karolinska (Sweden) were used: LBR2 Burkitt lymphoma, human breast cancer MDAMB-231, human pancreatic cancer PANC-1.
The cells were cultured at 37°C and 5% of СО2 in RPMI 1640 medium (PanEco, RF) containing 10% of fetal bovine serum (HyClone Laboratories, Logan, UK), inactivated at 56°C for 30 minutes, 2 mM of L- glutamine, 100 ug/ml of penicillin and 100 ug/ml of streptomycin sulfate (PanEco, RF). Light microscopy of cells was performed using 4 AxioVision system (Zeiss, Germany). Cell viability was determined by trypan blue dye exclusion (PanEco, Russia) in a Goryaev chamber.
Tumor cells, which reached the logarithmic growth phase were passaged into 96-well flat-bottomed microplates ("Costar") by 5·104- 6.5·104 cells per well, and preincubated for 24 h before adding test organisms under the conditions of СО2 5% and 37°С. The obtained cell cultures in a wide range progressively decreasing concentrations were added to cell culture wells (20 ul to 180 ul of cell suspension) and were co-incubated for 48 hours. Control wells were added of 0.9% sodium chloride solution in an adequate amount (20 ul). At the end of the incubation period, the number of living cells was determined in the wells of MTT by a colorimetric assay based on the ability of dehydrogenases of living cells to reduce 3-(4,5-dimethylthiazol-2-il)-2,5-diphenyltetrazolium bromide (MTT) to purple formazan crystals, soluble in dimethylsulfoxide. Optical absorption of dimethylsulfoxide colored solutions was measured on tablet photometer Multiskan MS (Labsystem, Finland) at λ = 540 nm.
Characterization and identification of LAB isolate from the gastrointestinal tract of man
To identify the strains of SK1, SK2, SK8, SK11 classification standard biochemical methods were used [49-52] using API20E, API 20 Ne, API Staphy Step and API (BioMerieux, France), in accordance with the recommendations of [53,54] which was followed by genetic identification by 16S rRNA sequence.
Phenotypic characterization of LAB strains
Classical phenotypic characteristics have been obtained in accordance with the phenotypic criteria. [55]. The morphology of cell strains was determined by biological microscope AxioScope A1 (Carl Zeiss, Germany), cell strains were tested using a Gram set for gram stain (Lab-Biomed, Moscow), or by catalyze reaction, the evolution of gas from glucose and growth capacity in MRS broth at 15°C for 7 days at 30°C, 37°C for 2 days.
Molecular identification and genotyping of LAB strains
Isolation of DNA microorganisms was performed using a set of reactants "PROBA-NK" to isolate DNA from biological material (DNA-Technology, Russia).
Agarose gel electrophoresis was performed using a chamber for horizontal electrophoresis SE-2 with a power supply "Elf-4" (Helicon, Russia). To prepare 2% of agarose 50 ml of single TBE buffer was added to 1 g of agarose, and mixed thoroughly. The obtained solution was placed in a microwave oven (for 2-5 minutes, depending on the capacity of the furnace, watching intensity of boiling suspension) or heated in a water bath for 15 minutes to completely dissolve the agarose. The molten agarose was cooled to 56 ° C and 5 ul of ethidium bromide (10 mg/ml) and was thoroughly stirred. The molten agarose and ethidium bromide were poured into the prepared pan. The thickness of the gel was 0.5-0.7 cm. After 30-40 minutes, the comb was removed. Finished gel was used immediately or stored in a single buffer in a refrigerator at 4 ° C. For electrophoresis, 2 ul of loading buffer and 10 ul of the reaction mixture were mixed in a separate tube. The mixture was added into the wells of the gel (the ratio of buffer/reaction mixture - 2/8). Molecular weight marker (100 bp) was added to one of the wells. Filled gel was placed in the electrophoresis chamber filled with buffer. The thickness of the buffer layer over the surface of the gel was about 2-3 mm. In constant voltage mode 100 B electrophoresis lasted 70-90 minutes. For the validation of the obtained results 100 bp of DNA-marker (SibEnzyme, Russia) was placed in one of the wells. Composition of buffer for storage: 10 mM of Tris HCl (pH 8,0); 1 mM of EDTA; 50 mM of NaCl.
Polymerase chain reaction was performed using the primers specially selected and synthesized by "Synthol" (Moscow). Temperature regime was selected, taking into account the length of the amplified fragment, and the length and composition of the primers used.
Isolation and purification of the PCR products were performed using GTG-agarose (Lonza Rockland, USA). Tubes with amplification products were consistently exhibited in a tripod, 1/5 of violet crystal colorant was added (MERCK, Germany) and stirred. They were taken from the oil layer by 20 microliters of samples and added to the wells of the gel, after which the chamber lid was closed. The DC electric current was connected to the camera, source parameters were exposed (voltage of 250V, current of 100 MA, 7 minutes duration), and then the correct polarity was switched on. The optimal electric field intensity in this case was 10 V/cm.
Upon completion of electrophoresis, the gel was placed on a transilluminator, placing the strips horizontally up the hole. Agarose gel lane was carefully cut out with a surgical blade, preliminarily treated with 70% of ethanol solution, containing the reaction product of required size PCR, trying to capture as less as possible of the gel and place it in a test tube with the appropriate number. Prior to cutting of each next segment the surgical scalpel was treated with 70% of ethanol in order to avoid contamination of the samples. Cut out strips of agarose gel containing the products of the reaction PCR were placed, inside of trimmed filter tips with a 200 microliters volume. Tubes were centrifuged for 5 minutes at maximum speed.
The quality of treatment and the number of the selected samples were evaluated. The horizontal agarose gel electrophoresis was repeated for this. Sequencing of the 16S RNA gene fragment of the isolated microorganisms was performed according to the Segnera method, using reagent kit "ABI Prism Big Dye Terminator Cycle Sequincing Ready Reaction Kits" (Applied biosystems, USA) using the GS Junior sequencer 454 (Roche, Switzerland). Purification of the reaction products was performed using the Sanger sorbent Sephadex G50 Superfine (Biosciences, Sweden).
DNA strains SK1, SK2, SK8, SK11 were amplified using primer set (Synthol Moscow) by the method of Weisburg et al. [56]. The obtained 16S rDNA - PCR products were purified by GFX Genomic Blood DNA Purification Kit (Amersham Biosciences) and DNA were used as template for a standard procedure of sequencing (Macrogen Inc, Seoul, Republic of Korea). Sequences edited to eliminate the PCR primer of binding sites and manually corrected Sequence Scanner 1.0 (Applied Biosystems), were compared with the available nucleotide GenBank database using the BLAST program. Generation of philogenetic trees was performed using Mega 6 software to align nucleotide sequences using the program BioEdit, to construct phylogenetic trees the nearest-neighbor method (Neiighbor-Joining NJ) of attachment was used [57]. Sequence of 16S Clostridium botulinum strain ELTDK 103 was used as the root. Stability of obtained phylogenetic trees was evaluated by bootstrap method.
Isolation and purification of bacteriocins
Isolation and purification of bacteriocins were performed by the method described in Russian patent №2492231 [58], i.e., the culture fluid is concentrated on hollow fibers, then dry NaCl is added, and stirred it on a rocker; suspension is centrifuged, the supernatant is adjusted to pH 3.0, the resulting suspension is centrifuged, the precipitate is added to water, then the precipitate is suspended and alcohol is added, incubated for 30 minutes at 0 ° C, centrifuged, the alcohol is removed from the solution by evaporation, water is added and activated charcoal, is centrifuged to remove adsorbed on carbon impurities, the aqueous solution is passed through the membrane.
Lipid bilayer structure and their oxidation products
Liposomes were prepared by injection method [59]. For this purpose, 5 ml of distilled water or an appropriate buffer solution with a constant vigorous rapid stirring was injected by syringe of 0.25 ml of the desired concentration of phospholipid in ethanol. Further, the liposomes were subjected to spontaneous and induced oxidation at a temperature of 37°C.
Lipid peroxidation products were labeled as: PR1 are carbonyl compounds, PR2 are substances which react with 2-thiobarbituric acid, PR3 are conjugated diene, PR4 is crotonaldehyde.
The concentration of carbonyl compounds was determined using N- (2,4-dinitrophenyl) hydrazine. The concentration of carbonyl compounds was determined using N- (2,4-dinitrophenyl) hydrazine. After 10 minutes the reaction mixture was added with 1 ml of 0.75 M NaOH. After 10 minutes, the absorbance of the reaction mixture in a spectrophotometer UV 1800 (Shimadzu, Japan) at a wavelength of 460 nm was measured.
The concentration of the substances in the suspension of liposomes reacting with 2-thiobarbituric acid was determined by the method described [60]. 0.5 ml of liposome suspension was added of 0.5 ml of 0.92 M of trichloroacetic acid and 1 ml of 49 mM 2-thiobarbituric acid, heated for 15 minutes in a water bath, centrifuged for 10 minutes at 3000 g, the supernatant is photometrically at wavelengths of 452 and 532 nm on a spectrophotometer UV 1800 (Shimadzu, Japan).
Determination of conjugated dienes in a suspension of liposomes was performed by the method proposed [61]. 0.2 ml of liposome suspension was extracted with 2 ml of heptane - 2-propanol (1:1) for 1 min with vigorous shaking, after which the system was added of 0.2 ml of water. After 10 min 0.2 mL was collected from the upper heptane phase and diluted with 0.2 ml of ethanol and photometrically at 232 nm using a spectrophotometer UV 1800 (Shimadzu, Japan).
The content of crotonaldehyde in the liposome suspension was determined by photometry of heptane extract at 220 nm.
Disc diffusion method
Antimicrobial activity of bifidobacteria strains was determined by agar-disc diffusion method [62,63] under anaerobic conditions. Test strain was plated on agar medium (RPA), lawn and turf imposed simultaneously on paper discs impregnated with metabolites of microorganisms (10 ul/disk). The antimicrobial activity was evaluated by measuring the inhibition zones against the microorganism of test culture [64]. Disc with a MRS medium was used as a control, disc with an antibiotic ciprofloxacin (from a standard set) was used as the reference preparation. The plates were incubated at 37°C for 24 hours. The results were taken into account on presence and size (in mm) of the transparent area with lack of microorganism growth around the disc.
The antioxidant properties of LAB
The effectiveness of the tested antioxidant activity of a chemical compound in each series of experiments and for each duration of liposomes oxidation in the model system of microorganisms, isolated from the intestinal tract was calculated according to the formula:
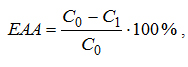
where С0 is the concentration of carbonyl compounds in the liposome suspension that does not contain test organisms (control), С1 is a concentration of carbonyl compounds in the liposome suspension containing the investigated microorganisms (experiment). If the calculated value >0, it is considered that the investigated microorganism inhibits lipid peroxidation, if <0, the bacteria in the study reinforces the lipid, i.e., it is a pro-oxidant.
LAB biocompatibility
The study of the biocompatibility was performed by co-culture method on solid MRS medium. Overnight culture, grown in a liquid medium and standardized by the turbidity standard was applied on the surface of a dense nutrient medium by bacteriological loop of 3 mm diameter. After drop soaking, departing 1-2 mm from the edge of the surface, the droplet of other test culture which was applied on the same medium in the same volume, spreading, covers about a half of the first drop. In the superimposed part cultures are developing in the mutual presence (co-cultivation), competing with each other. After drying of the second drop plates with crops turned upside down and incubated at 37-39°C in the air with an increased carbon dioxide content. Each experiment was repeated, changing the position of crops (to avoid the impact of successive layers of crop drops on the growth pattern in the co-cultivation area).
Controls were drops of the same culture, layered on each other as described above. Interpretation of results was performed after 24 and 48 hours after initiation of incubation. When one of the studied crops was stunting the relationship between them were regarded as antagonistic, and the cultures themselves belong to the category of bio-incompatibility. Cultures were considered as biocompatible in the case of a full "merger" spots, or enhance of the strains growth in the area of co-culture (mutualism, synergism, satellism). If one of the crops in the area of cocultivation "goes upstairs," inhibiting the growth of a second culture, regardless of the sequence of their application, then such an option was considered as a weak antagonism. The presence of well expressed zone of inhibition (growth retardation) of one culture by another peripheral spot of test culture was regarded as a sign of a strong antagonism.
Antiproliferation of colon cancer cells
MTT assay: MTT colorimetric method is based on ability of living cells to reduce dehydrogenase 3- (4,5-dimethylthiazol-2-yl) -2,5-diphenyltetrazolium bromide (MTT) to violet formazan crystals, soluble in dimethylsulfoxide. Optical absorption of colored solutions of dimethylsulfoxide was measured on tablet photometer Multiskan MS (Labsystem, Finland) at λ = 540 nm. Cytotoxicity of the tested cell cultures was evaluated according to the formula:
[% cell cytotoxicity = (1 - the optical absorption in the test sample/the optical absorption in control) * 100].
Trypan Blue exclusion assay: Cancer cells (106 cells/1 ml of cell suspension) were added to a 96-well plate. The suspension was incubated at 37 ° C for 24 h. Cell viability was examined using Trypan Blue exclusion:
[% cell viability = (live cell count/total cell count) *100].
The permeability of the cancer cells: Permeability of cancer cells was investigated in the chamber divided into two compartments by the permeable membrane filter [66]. Cancer cells were previously grown on this filter to a monolayer, and then transferred to a chamber in which both compartments contain DMEM medium. After 20 minutes of incubation for the initial establishment of equilibrium in the conditional inner "abdominal" compartment, containing monolayer dye-labeled Trypan Blue albumin (60 uM) was added there. Its concentration in the external "ex-abdominal" compartment was monitored by absorbance at 480 nm (Specord 10, Carl Zeiss, Germany). The flow of albumin across the surface of the monolayer was expressed in mole/(sec•cm²), permeability was represented as a coefficient (cm/sec), calculated based on the albumin concentration differences in the two compartments. To evaluate the influence of microbial metabolites on the permeability of cancer cells, they were grown with recent metabolites at concentrations ranging from 1-100 micrograms/ml for 24 hours and then transferred into the chamber.
Statistical analysis: All experiments were performed in triplicate. Data processing was carried out by the standard techniques of mathematical statistics. Differences between means are considered significant when the confidence interval is smaller than 5% (P £ 0.05).
Identification and molecular typing of LAB isolates
The strains were characterized as Gram-positive, slightly curved or branching, catalase-negative, immobile, not forming bacteria spores. They do not produce gas from glucose and grow well in MRS broth (pH 4.3-3.9) at a temperature of 37°C anaerobically that allows to classify them to the Actinobacteria class, Bifidobacteriaceae family [55]. Sequence analysis of 16s rDNA is a major step in the present study of isolates identification from the gastrointestinal tract of humans. BLAST analysis of the nucleotide sequences with the available GenBank database, showed almost 100% similarity with strains of SK Bifidobacterium kinds (Table 1 and Figure 1).
Table 1. Identification of the isolated Bifidobacterium from gastro-intestinal tract of man by 16S rDNA sequence analysis.
Strain |
Identified as: |
Probability [%] based on the BLAST analysis of the sequences |
Identified BLAST analysis as: |
SK1 |
Bifidobacterium |
100% |
B.bifidum |
SK2 |
Bifidobacterium |
100% |
B. breve |
SK8 |
Bifidobacterium |
100% |
B. longum |
SK11 |
Bifidobacterium |
99% |
B. adolescentis |
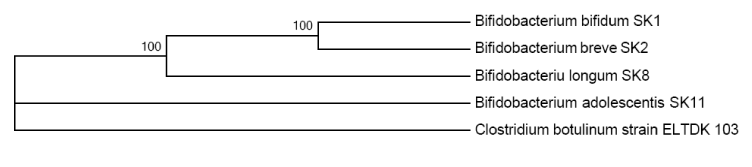
Figure 1. Phylogenetic tree based on Bifidobacteria 16s rDNA of sequences of the four SK, isolated from the gastrointestinal tract of humans
In vitro study of antioxidant activity of bifidobacteria
The antioxidant effect of micro-organisms was evaluated in the oxidation of liposomes over time. In the course of reactions of lipid peroxidation product formation was studied: PR1 are carbonyl compounds, PR2 are substances which react with 2-thiobarbituric acid, PR3 are conjugated dienes, PR4 is crotonaldehyde. The results are presented in Figure 2. All the studied strains of microorganisms reduce the amount of liposomes in suspensions subjected to induced oxidation, lipid peroxidation of products, hence the mechanism of antioxidant action of probiotic microorganisms lies in inhibition of lipid peroxidation metabolites that make up cell membranes. This is likely due to binding of the free radicals and the inhibition of the lipoxygenase enzyme.
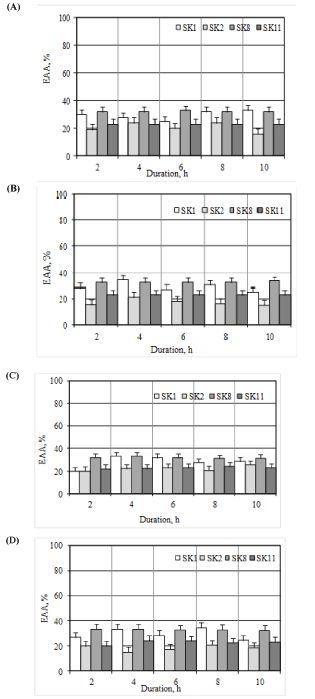
Figure 2. The effectiveness of the antioxidant activity of a product relative to strains of LAB (A) PR1, (B) PR2, (C) PR3, (D) PR4 from duration of oxidation
In vitro study of the antimicrobial activity of bifidobacteria
Antimicrobial activity of the microorganisms isolated from the intestinal tract of humans was studied by inhibitory action on test cultures. Results are presented in Figure 3. Metabolites produced by microorganisms of Bifidobacterium genus, exhibit antimicrobial activity against the test strains of E. coli В-6954, Staphylococcus aureus ATCC 25923, Salmonella enterica ATCC 14028, Klebsiella pneumoniae В-7001. The greatest breadth of the antimicrobial activity was demonstrated by SK2 strain, which have an inhibitory effect also on Clostridium tyrobutyricum LMG.
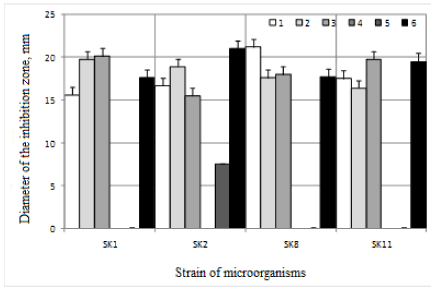
Figure 3. The antimicrobial properties of the metabolites synthesized by microorganisms isolated from the gastrointestinal tract of humans: 1 is E. coli B-6954; 2 is Staphylococcus aureus ATCC 25923; 3 is Salmonella enterica ATCC 14028; 4 is Listeria innocua LMG; 5 is Clostridium tyrobutyricum LMG; 6 is Klebsiella pneumoniae B-7001
Identification of bifidobacteria metabolites
In the process of life activity, lactic acid bacteria form alcohols, acids, esters, vitamins, antibacterial agents, and others. Antibiotics or bacteriocins have antimicrobial qualities, distinctive feature of latter is their safety. An important component of the study was to prove affiliation of metabolites of the isolated bifidobacteria to bacteriocins. For this purpose the physical, chemical and biological properties were studied. The results are shown in Table 2.
Table 2. Physical and chemical properties of metabolites synthesized by isolated Bifidobacterium
Strain |
Properties of metabolites |
Molecular mass, kDa |
Solubility |
Insolubility |
Bifidobacterium SK1 |
38.0 |
in ethyl acetate, the aqueous acetonitrile |
in water, ethanol |
Bifidobacterium SK2 |
35.5 |
chloroform, acetonitrile |
in water, n-butanol |
Bifidobacterium SK8 |
38.8 |
in ethyl acetate, the aqueous acetonitrile |
in water, ethanol |
Bifidobacterium SK11 |
39.0 |
in ethyl acetate, the aqueous acetonitrile |
in water, ethanol |
Typical quality of bacteriocins is defined by the restriction of species and strains activity, particularly of one type of subspecies. Immunity of producer strain to its own bacteriocin was determined using producing strain as indiatornoy culture. The results obtained are shown in Table 3. All SK strains, isolated from the gastrointestinal tract of human are resistant to both inhibitors own and mutually resistant to synthesized substances of one kind, indicating the affinity of substances produced and their bacteriocin-like nature.
Table 3. Study of the cross-sensitivity of microorganisms strains, producing bacteriocins, by the diameter of the inhibition zone (mm)
Strain |
SK1 |
SK2 |
SK8 |
SK11 |
L1 |
L2 |
L3 |
L4 |
L5 |
L6 |
L7 |
L8 |
S9 |
E10 |
Diameter of the inhibition zone, mm |
SK1 |
0 |
0 |
0 |
0 |
5.4 |
4.2 |
3.3 |
5.6 |
7.0 |
4.4 |
2.1 |
1.5 |
3.0 |
4.1 |
SK2 |
0 |
0 |
0 |
0 |
6.0 |
4.5 |
5.3 |
4.8 |
3.9 |
4.0 |
3.5 |
2.2 |
3.5 |
4.6 |
SK8 |
0 |
0 |
0 |
0 |
7.1 |
4.5 |
3.8 |
5.7 |
3.0 |
4.2 |
5.4 |
3.7 |
5.0 |
5.1 |
SK11 |
0 |
0 |
0 |
0 |
2.4 |
4.0 |
3.6 |
3.5 |
6.0 |
3.2 |
4.9 |
3.6 |
5.5 |
3.6 |
L1 |
4.7 |
3.6 |
2.9 |
3.8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1.7 |
2.8 |
L2 |
2.9 |
3.2 |
4.4 |
3.6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
3.0 |
4.1 |
L3 |
1.5 |
1.7 |
3.5 |
4.2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2.6 |
2.9 |
L4 |
1,8 |
1.6 |
1.5 |
2.4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2.2 |
3.4 |
L5 |
5.3 |
7.0 |
3.6 |
4.7 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1.9 |
2.5 |
L6 |
4.5 |
4.7 |
3.6 |
5.0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2.8 |
3.2 |
L7 |
3.3 |
2.7 |
1.6 |
4.3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1.8 |
2.2 |
L8 |
2.7 |
4.4 |
2.2 |
3.6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
4.3 |
5.1 |
S9 |
4.5 |
3.9 |
5.1 |
6.0 |
3.2 |
2.7 |
3.1 |
4.7 |
2.5 |
3.6 |
4.0 |
3.5 |
0 |
4.7 |
E10 |
3.0 |
4.2 |
1.9 |
2.5 |
2.9 |
3.4 |
5.1 |
6.3 |
4.2 |
4.0 |
3.6 |
2.8 |
2.9 |
0 |
* L1 - L. plantarum, L2 - L. acidophilus, L3 - L. rhamnosus, L4 - L. paracasei, L5 - L. fermentum, L6 - L. salivarius, L7 - L. casei, L8 - L. reuteri, S9 - St. agalactiae, E10 - E. faecium
Comparison of the data obtained (Table 2 and 3) and references data [67-71] indicates that the identified metabolites of microorganisms isolated from the intestinal tract of humans, are the bacteriocins.
In vitro study of antitumor activity of bifidobacteria
To examine the possible inhibitory effect of bifidobacteria bacteriocins, isolated from the gastro-intestinal tract of humans, on cancer cell cultures, the strain of Bifidobacterium breve was selected, having the highest antimicrobial activity.
Influence of microbial metabolites Bifidobacterium breve on the permeability of cell membranes was studied on cell lines of Burkitt's lymphoma LBR2, human breast cancer MDAMB-231 and human pancreatic cancer PANC-1. Determination results of cancer cells survival and the tumor cell permeability in the presence of Bifidobacterium breve microorganisms metabolites are shown in Figures 4 and 5, respectively. Bifidobacterium breve metabolites reduce survival of cancer cells of Burkitt lymphoma LBR2, human breast cancer MDAMB-231 and human pancreatic cancer PANC-1. Within 24 hours of cancer cell growth in the presence of Bifidobacterium breve metabolites the growth inhibition does not exceed 40.0%, while after 48 hours such inhibition is 67.6-71.1%. Moreover, the processing of cancer cells by Bifidobacterium breve metabolites causes an increase in permeability of the membranes for albumin. Increased permeability of the cell membrane as compared to untreated cells is 10.0-30.0%.
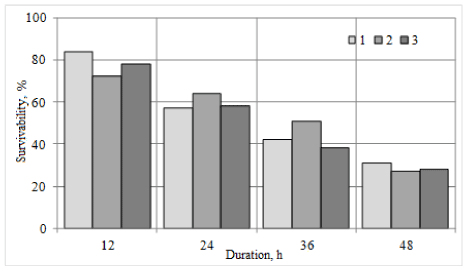
Figure 4. Survival of cancer cells under the influence of Bifidobacterium breve metabolites: 1 - LBR2 Burkitt lymphoma cells; 2 - Cells of human breast cancer MDAMB-231; 3 - cells of human pancreatic cancer PANC-1. Cell survival is taken for 100%, growing without Bifidobacterium breve metabolites
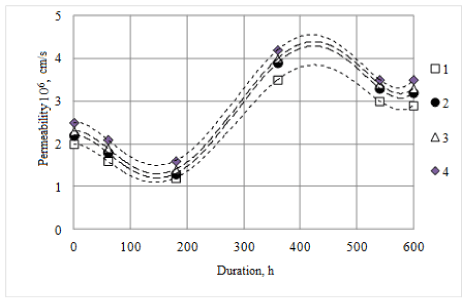
Figure 5. Dynamics of permeability changes for cancer cells albumin monolayer treated with Bifidobacterium breve metabolites: 1 - untreated cells; Burkitt's lymphoma cells LBR2 treated with metabolites of Bifidobacterium breve; 2 - cells of human breast cancer MDAMB-231 treated with metabolites of Bifidobacterium breve; 3 - cells of human pancreatic cancer PANC-1 treated with metabolites of Bifidobacterium breve
The study shows the detection results of bifidobacteria qualities with antimicrobial and antioxidant activity, isolated from the gastrointestinal tract of humans. A variety of representatives of the gastrointestinal tract of humans microbiota, combined with the distinctive features of his nutrition and the environment make the relevance of research of the human microcosm, including representing by lactic acid bacteria. Features of the variety and quality of lactic acid bacteria extend the capabilities of their use for human benefit and health. The set of methods (sequencing, combined with phylogenetic analysis) allowed identifying isolates of SK1, SK2, SK8 and SK11 (Table 1 and Figure 1) as bifidobacteria with sufficient confidence probability within the limits of specific accessory (the probability of error does not exceed 0.05, which corresponds to statistical error). The available information [20,72,73] on biochemical research methods (API analysis using APILAB software), is not always with a sufficient accuracy relates to the appropriate test organisms and requires the final stage of the use of phylogenetic analysis, which we chose as the main on this phase of the study. Analysis of 16s rDNA sequences is uniquely identified SK1, SK2, SK8, SK11 strains as the representatives of the species Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium adolescentis, respectively (for an unambiguous determination of the accuracy type of the similarity to consensus sequence of 16s rDNA is required [74]). BLAST-analysis showed a high similarity with the types of Bifidobacterium bifidum strain (L22), Bifidobacterium breve strain (BB4GZU), Bifidobacterium longum strain (ATCC 15707), Bifidobacterium adolescentis strain (Ru424).
Probiotics have a positive impact on the immune system, as a gastro-intestinal flora leads to reducing the impact of mutagens and carcinogens. In vitro experiments demonstrated that the investigated microorganism strains reduce the amount of lipid peroxidation, therefore, the mechanism of antioxidant action of probiotic microorganisms is inhibiting lipid peroxidation metabolites that make up cell membranes. The antioxidant abilities of the new dedicated SK1, SK2, SK8, SK11 LAB strains, demonstrated in this study, confirm already published data on bifidobacteria [75-77], and promote the opportunities for their use as basis products for rehabilitation of patients after chemotherapy or radiation.Like the other lactobacteria, bifidobacteria are used in the treatment and prevention of a wide spectrum of diseases ranging from gastrointestinal tract and finishing with cosmetology [78-83].
Four isolated Bifidobacterium strains, designated as SK1, SK2, SK8, SK11 demonstrated a broad spectrum of antimicrobial activity. MRS was used as a medium in our in vitro studies, and traditional pathogens, causing both active spoilage of food, and leading to serious human diseases as test cultures [84,85].
The conducted experiments using the agar disk diffusion method of the test bifidobacteria confirmed an inhibitory effect on the growth of certain rot microorganisms. Strains of Bifidobacterium SK1, SK2, SK8, SK11 were entirely inhibited by E. coli B-6954, Staphylococcus aureus ATCC 25923, Salmonella enterica ATCC 14028, Klebsiella pneumoniae B-7001, remained tolerant to Listeria innocua LMG. One of the isolated strains of Bifidobacterium breve SK2 also inhibited the growth of Clostridium tyrobutyricum LMG [86,87].
Bacteriocins of lactic acid bacteria (LAB) are a variety of proteins or peptides which have antimicrobial properties, have application in the food industry in the form of bio preservatives and medicine as potential alternatives to antibiotics [46,47,63-71,86,87]. There is a constant screening of LAB, new producers of bacteriocins, their properties and characteristics attract constant attention. The antimicrobial, antioxidant, antifungal activity of bacteriocins are well known [12-15,17-23,47,48,64,69,75-77,88,89]. But perhaps there are also qualities that were not studied by a human. We have attempted to identify the inhibitory action is not only against similar to themselves bacteria, but also against cancer cell cultures. Therefore, it was important for us to confirm that the metabolites of bifidobacteria isolated from the gastro-intestinal tract of man are metabolites, and after setting the fact to obtain confirmation of inhibitory effect on cancer cells.
Practically there are no data on the inhibitory effects of lactic acid bacteria and their metabolites on the cancer cell cultures in the scientific literature [28,29,34,36,38,39,90-92]. Partial inhibition of cancer cells growth by bacteriocins of Bifidobacterium breve strain was already observed after 12 hours after treatment with metabolites and, after 36 hours after treatment no more than half of the cell cultures remained. Also, the membrane permeability of cancer cells of the species is increasing, immediately after the treatment of 10 to 25%, after 60 hours from 12.5 to 31.25%, depending on the types of cancer cells. The result is more than just positive, opening the possibility of new treatment technologies of this disease.
The experiments have found that the treatment of one isolated metabolites strains SK2 leads to decreased survival and increased membrane permeability of cancer cells of Burkitt lymphoma LBR2, human breast MDAMB-231 and human pancreatic cancer PANC-1, inhibiting the growth of P. claviforme (Figures 4 and 5).
In this study, four is2021 Copyright OAT. All rights reservfrom the gastrointestinal tract of human, have been identified as Bifidobacterium and characterized as a promising culture with antioxidant, antimicrobial and antitumor activity (Bifidobacterium breve). The antioxidant activity of our new isolates identified as Bifidobacterium, involves their potential use in a variety of food technology, including products for the rehabilitation of patients in the period of remission. The obtained results of studying the possible use of lactic acid bacteria metabolites in cancer therapy and prevention are the initial and insufficient. There is no complete picture of the mechanisms’ effects on cancer cell cultures and it requires further study, both in terms of the expansion of a number of microorganisms, and in terms of the study of their properties.
Financial support for this research was provided by the Ministry of Education and Science of the Russian Federation in the framework of the state contract No.14.586.21.0002 and research partner of the Department of Microbiology, Tumor and Cell Biology, Karolinska Institute (Stockholm, Sweden).
- Murray CJ, Lopez AD (1996) The Global Burden of Disease: a comprehensive assessment of mortality and disability from diseases, injuries and risk factors in 1990 and projected to 2020. Boston: Harvard School of Public Health, on behalf of the World Health Organization and the World Bank, 1996.
- Hjartåker A, Weiderpass E, Bray FI (2008) Cancer Mortality. In: Heggenhougen K, Quah S (Eds.), International Encyclopedia of Public Health, Academic Press 452–464.
- Anttila A, Lönnberg S, Ponti A, Suonio E, Villain P, et al. (2015) Towards better implementation of cancer screening in Europe through improved monitoring and evaluation and greater engagement of cancer registries. Eur J Cancer 51: 241–251. [Crossref]
- Von Karsa L, Dean PB, Arrossi S, Sankaranarayanan R (2014) Screening – principles. In: Stewart BW, Wild CP, editors. World cancer report 2014. International Agency for Research on Cancer: Lyon 322–329.
- Eslick GD (2010) Infectious causes of esophageal cancer. Infect Dis Clin North Am 24: 845-852. [Crossref]
- Porta C, Riboldi E, Sica A (2011) Mechanisms linking pathogens-associated inflammation and cancer. Cancer Lett 305: 250-262. [Crossref]
- De Martel C, Plummer M, Franceschi S (2014) Infections causing cancers: world burden and potential for prevention. Public Health Forum 22: 12e1-12e4.
- Velázquez E, Peix A, Gómez-Alonso A (2011) Microorganisms and cancer: scientific evidence and new hypotheses. Cir Esp 89: 136-144. [Crossref]
- Sarid R, Gao SJ (2011) Viruses and human cancer: from detection to causality. Cancer Lett 305: 218-227. [Crossref]
- Read SA, Douglas MW (2014) Virus induced inflammation and cancer development. Cancer Lett 345: 174-181. [Crossref]
- Irigaray P, Newby JA, Clapp R, Hardell L, Howard V, et al. (2007) Lifestyle-related factors and environmental agents causing cancer: an overview. Biomed Pharmacother 61: 640-658. [Crossref]
- Bianchini A (2015) Lactic acid bacteria as antifungal agents. In: Advances in Fermented Foods and Beverages, Elsevier Science, Woodhead Publishing 333-353.
- O'Bryan CA, Crandall PG, Ricke SC, Ndahetuye JB (2015) Lactic acid bacteria (LAB) as antimicrobials in food products: Analytical methods and applications. In: Handbook of Natural Antimicrobials for Food Safety and Quality, Elsevier Science, Woodhead Publishing 137-151.
- Iranmanesh M, Ezzatpanah H, Mojgani N (2014) Antibacterial activity and cholesterol assimilation of lactic acid bacteria isolated from traditional Iranian dairy products. LWT-Food Sci Technol 58: 355-359.
- Inglin RC, Stevens MJ, Meile L, Lacroix C, Meile L (2015) High-throughput screening assays for antibacterial and antifungal activities of Lactobacillus species. J Microbiol Methods 114: 26-29. [Crossref]
- Aguirre L, Hebert EM, Garro MS, de Giori GS (2014) Proteolytic activity of Lactobacillus strains on soybean proteins. LWT - Food Sci Technol 59: 780-785.
- Cizeikiene D, Juodeikiene G, Paskevicius A, Bartkiene E (2013) Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 31: 539-545.
- García-Ruiz A, Requena T, Peláez C, Bartolomé B, Moreno-Arribas MV, et al. (2013) Antimicrobial activity of lacticin 3147 against oenological lactic acid bacteria. Combined effect with other antimicrobial agents. Food Control 32: 477-483.
- Shori AB (2013) Antioxidant activity and viability of lactic acid bacteria in soybean-yogurt made from cow and camel milk. J Taibah Univ Sci 7: 202-208.
- Tropcheva R, Nikolova D2, Evstatieva Y2, Danova S3 (2014) Antifungal activity and identification of Lactobacilli, isolated from traditional dairy product "katak". Anaerobe 28: 78-84. [Crossref]
- Coloretti F, Carri S, Armaforte E, Chiavari C, Grazia L, et al. (2007) Antifungal activity of lactobacilli isolated from salami. FEMS Microbiol Lett 271: 245-250. [Crossref]
- Magnusson J, Ström K, Roos S, Sjögren J, Schnürer J (2003) Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol Lett 219: 129-135. [Crossref]
- Schnürer J, Magnusson J (2005) Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci Technol 16: 70-8.
- Burns AJ, Rowland IR (2000) Anti-carcinogenicity of probiotics and prebiotics. Curr Issues Intest Microbiol 1: 13-24. [Crossref]
- Russell DA, Ross RP, Fitzgerald GF, Stanton C (2011) Metabolic activities and probiotic potential of bifidobacteria. Int J Food Microbiol 149: 88-105. [Crossref]
- Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, et al. (2014) Probiotic potential and biotherapeutic effects of newly isolated vaginal Lactobacillus acidophilus 36YL strain on cancer cells. Anaerobe 28: 29-36. [Crossref]
- Arici M, Bilgin B, Sagdic O, Ozdemir C (2004) Some characteristics of Lactobacillus isolates from infant faeces. Food Microbiol 21: 19-24.
- Gonet-Surowka AK, Strus M, Heczko PB (2007) Influence of Lactobacilli probiotic strains on apoptosis of colon cancer cells lines. Int J Antimicrob Agents 29: 343–344.
- Thirabunyanon M, Boonprasom P, Niamsup P (2009) Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol Lett 31: 571-576. [Crossref]
- Zhu Y, Michelle Luo T, Jobin C, Young HA (2011) Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett 309: 119-127. [Crossref]
- Fuller R (1991) Probiotics in human medicine. Gut 32: 439-442. [Crossref]
- Goldin BR, Gorbach SL (2008) Clinical indications for probiotics: an overview. Clin Infect Dis 46: S96-100. [Crossref]
- Darsanaki RK, Issazadeh K, Pahlaviani MRMK, Aliabadi MA (2012) Antimutagenic activity of Lactobacillus Isolated from fresh vegetables against Sodium Azide and 2-Nitrofluorene. J Pure Appl Microbiol 6: 1677-1682.
- Commane D, Hughes R, Shortt C, Rowland I (2005) The potential mechanisms involved in the anti-carcinogenic action of probiotics. Mutat Res 591: 276-289. [Crossref]
- Maragkoudakis PA, Zoumpopoulor G, Miaris C, Kalantzopoulos G, Pot B, et al. (2006) Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J 16: 189–199.
- de Moreno de LeBlanc A, Matar C, Perdigón G (2007) The application of probiotics in cancer. Br J Nutr 98 Suppl 1: S105-S110. [Crossref]
- Pennacchia C, Ercolini D, Blaiotta G, Pepe O, Mauriello G, et al. (2004) Selection of Lactobacillus strains from fermented sausages for their potential use as probiotics. Meat Sci 67: 309-317. [Crossref]
- Rafter J (2003) Probiotics and colon cancer. Best Pract Res Clin Gastroenterol 17: 849-859. [Crossref]
- Saikali J, Picard C, Freitas M, Holt P (2004) Fermented milks, probiotic cultures, and colon cancer. Nutr Cancer 49: 14-24. [Crossref]
- Gaggia F, Di Gioia D, Baffoni L, Biavati B (2011) The role of protective and probiotic cultures in food and feed and their impact in food safety. Trends Food Sci Technol 22: S58-66.
- Oliveira PM, Zannini E, Arendt EK (2014) Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: from crop farming to cereal products. Food Microbiol 37: 78-95. [Crossref]
- Plockova M, Stiles J, Chumchalova J, Halfarova R (2001) Control of mould growth by Lactobacillus rhamnosus VT1 and Lactobacillus reuteri CCM 3625 on milk agar plates. Czech J Food Sci 19: 46-50.
- Valerio F, Lavermicocca P, Pascale M, Visconti A (2004) Production of phenyllactic acid by lactic acid bacteria: an approach to the selection of strains contributing to food quality and preservation. FEMS Microbiol Lett 233: 289-295. [Crossref]
- Dalié DKD, Deschamps AM, Richard-Forget F (2010) Lactic acid bacteria - potential for control of mould growth and mycotoxins: a review. Food Control 21(4): 370-80.
- Delavenne E, Ismail R, Pawtowski A, Mounier J, Barbier G, et al. (2012) Assessment of lactobacilli strains as yogurt bioprotective cultures. Food Control 30: 206-13.
- Lacroix C (2011) Protective cultures, antimicrobial metabolites and bacteriophages for food and beverage biopreservation. Elsevier Science, Woodhead Publishing.
- Ravi V, Prabhu M, Subramanyam D (2011) Isolation of bacteriocin producing bacteria from mango pulp and its antimicrobial activity. J Microbiol Biotech Res 1: 54-63
- De Muynck C1, Leroy AI, De Maeseneire S, Arnaut F, Soetaert W, et al. (2004) Potential of selected lactic acid bacteria to produce food compatible antifungal metabolites. Microbiol Res 159: 339-346. [Crossref]
- Colwell RR, Grigorova R (1987) Methods in Microbiology. Volume 19: Current Methods for Classification and Identification of Microorganisms. London: Academic Press, Elsevier Science.
- Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH (1995) Manual of Clinical Microbiology. 6th ed. Washington, ASM Press, DC.
- Murray PR, Rosenthal KS, Pfaller MA (2013) Medical Microbiology. Saunders/Elsevier.
- Jorgensen JH (1993) Selection criteria for an antimicrobial susceptibility testing system. J Clin Microbiol 31: 2841-2844. [Crossref]
- Jorgensen JH, Tunridge JD, Washington JA (1999) Antibacterial Susceptibility Tests: Dilution and Disk Diffusion Methods. In: Manual of Clinical Microbiology. 7th eds. Washington, DC: American Society for Microbiology.
- Kandler O, Weiss N (1986) Regular, non-sporing gram positive rods. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey's manual of systematic bacteriology. Baltimore: Williams & Wilkins p. 1208-34.
- Bergey’s Manual of Systematic Bacteriology (1985) Book Review Int J of Syst Bact.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697-703. [Crossref]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425. [Crossref]
- Суровцев ВИ, Борзенков ВМ, Хатюшин ЮИ, Веревкин ВВ, Красильникова ВМ (2013) RU Patent №2492231.
- Batzri S, Korn ED (1973) Single bilayer liposomes prepared without sonication. Biochim Biophys Acta 298: 1015-1019. [Crossref]
- Esterbauer H, Cheeseman KH (1990) Determination of aldehydic lipid peroxidation products: malonaldehyde and 4-hydroxynonenal. Methods Enzymol 186: 407-421. [Crossref]
- Ушкалова ВН, Иоанидис НВ, Кадочникова ТД, Деева ЗМ (1993) Контроль перекисного окисления липидов. Новосибирск: изд-во НГУ.
- Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45: 493-496. [Crossref]
- Kimura H, Sashihara T, Matsusaki H, Sonomoto K, Ishizaki A (1998) Novel bacteriocin of Pediococcus ISK-1 isolated from well-aged bed of fermented rice bran. Ann N Y Acad Sci 864: 345-348. [Crossref]
- Shillinger U, Lücke FK. Antibacterial activity of Lactobacillus sake isolated from meat. J Appl Microbiol 1989; 70: 473-478.
- Eloff JN (1998) A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med 64: 711-713. [Crossref]
- Noll T, Wozniak G, McCarson K, Hajimohammad A, Metzner HJ, et al. (1999) Effect of factor XIII on endothelial barrier function. J Exp Med 189: 1373-1382. [Crossref]
- Ohland CL, Macnaughton WK (2010) Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol 298: G807-819. [Crossref]
- Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP (2013) Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv 31: 482-488. [Crossref]
- Poltavska OA, Kovalenko NK (2012) Antimicrobial activity of bifidobacterial bacteriocin-like substances. Mikrobiol Z 74: 32-42. [Crossref]
- Renye JA Jr, Somkuti GA (2013) BlpC-regulated bacteriocin production in Streptococcus thermophilus. Biotechnol Lett 35: 407-412. [Crossref]
- Rossi F, Marzotto M, Cremonese S, Rizzotti L, Torriani S (2013) Diversity of Streptococcus thermophilus in bacteriocin production; inhibitory spectrum and occurrence of thermophilin genes. Food Microbiol 35: 27-33. [Crossref]
- Wang J, Chen X, Liu W, Yang M, Zhang H (2008) Identification of Lactobacillus from koumiss by conventional and molecular methods. Eur Food Res Technol 227(5): 1555-1561.
- Delétoile A, Passet V, Aires J, Chambaud I, Butel MJ, et al. (2010) Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res Microbiol 161: 82-90. [Crossref]
- Bevilacqua L, Ovidi M, Di Mattia E, Trovatelli LD, Canganella F (2003) Screening of Bifidobacterium strains isolated from human faeces for antagonistic activities against potentially bacterial pathogens. Microbiol Res 158: 179-185. [Crossref]
- Shen Q, Zhang B, Xu R, Wang Y, Ding X, Li P (2010) Antioxidant activity in vitro of the selenium-contained protein from the Se-enriched Bifidobacterium animalis Anaerobe 16: 380-386.
- Xu R1, Shang N, Li P (2011) In vitro and in vivo antioxidant activity of exopolysaccharide fractions from Bifidobacterium animalis Anaerobe 17: 226-231. [Crossref]
- Chang OK, Seol KH, Jeong SG, Oh MH, Park BY, et al. (2013) Casein hydrolysis by Bifidobacterium longum KACC91563 and antioxidant activities of peptides derived therefrom. J Dairy Sci 96: 5544-5555. [Crossref]
- Watson RR, Preedy VR, editors (2010) Bioactive Foods in Promoting Health: Probiotics and Prebiotics. London: Academic Press.
- Barrons R, Tassone D (2008) Use of Lactobacillus probiotics for bacterial genitourinary infections in women: a review. Clin Ther 30: 453-468. [Crossref]
- Abdin AA, Saeid EM (2008) An experimental study on ulcerative colitis as a potential target for probiotic therapy by Lactobacillus acidophilus with or without "olsalazine". J Crohns Colitis 2: 296-303. [Crossref]
- Patel AR, Shah NP, Prajapati JB (2014) Immunomodulatory effects of probiotics in the treatment of human immunodeficiency virus (HIV) infection. Biomed Prev Nutr 4: 81-84.
- Hamilton-Miller JM (2003) The role of probiotics in the treatment and prevention of Helicobacter pylori infection. Int J Antimicrob Agents 22: 360-366. [Crossref]
- Falagas M, Betsi GI, Athanasiou S (2007) Probiotics for the treatment of women with bacterial vaginosis. Clin Microbiol Infect 13: 657-664. [Crossref]
- Filtenborg O, Frisvad JC, Thrane U (1996) Moulds in food spoilage. Int J Food Microbiol 33: 85-102. [Crossref]
- Delavenne E, Ismail R, Pawtowski A, Mounier J, Barbier G, et al. (2012) Assessment of lactobacilli strains as yogurt bioprotective cultures. Food Control 30: 206-13.
- Liu G, Ren L, Song Z, Wang C, Sun B (2015) Purification and characteristics of bifidocin A, a novel bacteriocin produced by Bifidobacterium animalis BB04 from centenarians' intestine. Food Control 50: 889-895.
- Gänzle MG, Hertel C, van der Vossen JM, Hammes WP (1999) Effect of bacteriocin-producing lactobacilli on the survival of Escherichia coli and Listeria in a dynamic model of the stomach and the small intestine. Int J Food Microbiol 48: 21-35.[Crossref]
- Lahtinen SJ, Jalonen L, Ouwehand AC, Salminen SJ (2007) Specific Bifidobacterium strains isolated from elderly subjects inhibit growth of Staphylococcus aureus. Int J Food Microbiol 117: 125-128. [Crossref]
- Cheikhyoussef A, Pogori N, Chen W, Zhang H (2008) Antimicrobial proteinaceous compounds obtained from bifidobacteria: from production to their application. Int J Food Microbiol 125: 215-222. [Crossref]
- Kittakoop P (2015) Anticancer Drugs and Potential Anticancer Leads Inspired by Natural Products. In: Studies in Natural Products Chemistry 44: 251–307.
- Collado MC, González A, González R, Hernández M, Ferrús MA, et al. (2005) Antimicrobial peptides are among the antagonistic metabolites produced by Bifidobacterium against Helicobacter pylori. Int J Antimicrob Agents 25: 385-391. [Crossref]
- Miki K, Urita Y, Ishikawa F, Iino T, Shibahara-Sone H, et al. (2007) Effect of Bifidobacterium bifidum fermented milk on Helicobacter pylori and serum pepsinogen levels in humans. J Dairy Sci 90: 2630-2640. [Crossref]