Abstract
The origin of life on earth demands a prerequisite for abiogenic evolution of RNA which is believed to be highly destructive on the earth covered by boiling ocean. I have proposed a mechanism of evolution of RNA as a left-handed single-stranded helical lattice with a trend of condensation and dehydration of organic compounds under a condition of high pressure and high temperature in the mantle of the earth. All organic compounds and energy-rich polyphosphates were synthesized from a large amount of methane hydrate, inorganic phosphate and oxygen radicals that were produced from ferrous oxides by disintegration of uranium. The six-fold symmetrical structure of the lattice allowed to stack a lot of energy-rich compounds such as adenosine triphosphate and phosphor-lipids which were required for formation of cell membrane at the later stage. More importantly, the cavity formed inside the lattice allowed polymerization of small L-amino acids such as glycine, alanine, proline and serine that were required to protect RNAs from hydrolysis. The 6.5-fold symmetrical lattice allowed a little more larger L-amino acids to incorporate the protective proteins and formation of α–helices, but insertion of larger amino acids such as arginine and methionine were rather difficult. However, in the next stage of evolution of the lattice formation of left-handed single-stranded RNA, an 8 fold symmetrical lattice seems to incorporate arginines and the other larger amino acids, and also a dimeric relationship between the two neighbor strands suggests a possibility of aminoacyl-tRNA synthetase to evolve because some of the aminoacyl-tRNA synthetases were formed to be in dimeric form.
Introduction
I have discussed how first life was born on earth in the hot mantle as a lattice of left-handed single-stranded helices under a severe condition of high pressure and high temperature [1]. The hypothesis how first life was born on earth is deeply related with a basic question how energy-rich substances were produced in abiogenic era of the earth. Key words are oxygen radicals. At the second stage of history of earth, if we follow the explanation of Nagano (2017), disintegration of uranium atoms generated a huge amount of thermal energy, and ferrous oxide created a lot of oxygen radicals. Oxygen radicals reacted with inorganic phosphates to produce energy-rich polyphosphates, resulting in accumulation of ATP, GTP, UTP, etc. Inositol triphosphate, which plays a big role in calcium control in a cell, is also one of them. At the first stage in the history of earth, it contained a lot of methane hydrate, which was changed into a various pentoses and hexoses [1,2]. Among those sugars, only 5’-phospho-D-ribose could grow up to a big world as left-handed single-stranded helices, forming an infinite size of layers. Uracil-Uracil base pairs must have been very important to connect with each other and support the neighboring layers. The reaction between phosphoglyceride and fatty acids would have formed a phospholipid making a big membrane inside the mantle containing a chaotic mixture of pentoses and hexoses, as well as ammonia, L- and D-amino acids. According to the order of gravity, phosphorus atom containing substances had a tendency to concentrate in the center of the mantle. Among various pentoses and hexoses, 5’-phospho-D-ribose could only have a chance to polymerize under a condition of high pressure and temperature. It is, however, a matter of delicate balance between the RNA layers and the surrounding rocks. If the temperature is too high, the most volatile elements would have been evaporated except carbon atoms and would produce diamond under a suitable condition of high pressure. In such a suitable condition of high pressure and temperature, the driving force of polymerization could have shifted from oxygen radical reaction to condensation and dehydration due to the environmental condition. Evolution of polypeptides as a supporting substance was also very important.
Accumulation of small L-amino acids in the cavity of RNA layers
Figure 1 shows a stereoview of three 6-fold left-handed single- stranded helices of 5’-phospho-D-ribose connected with each other by Uracil-Uracil base pairs. Since the base stacking distance of the layer is 7.2A, various nucleotide triphosphates can be intercalated as shown in Figure 2, which is an example for ATP drawn in green. Polymerization of small L-amino acids growing up in the hole of cavity, even if it is not a right-handed α–helix. It is important to know that L-amino acids are more favorable to be incorporated than D-amino acids under such a stereochemical condition. The example shown in Figure 3 is Gly-Pro-Gly-Asp-Ala-Ala-Ser-Val. In the primordial period of such a protein synthesis, some amount of D-amino acids could be involved, but such a case could result in infertility and digestion at the outside of the mantle. Another problem is whether ATP could take part in the protein synthesis or not. It is clear that aminoacyl-tRNA synthetase in the later stage require ATP. However, when temperature and pressure are too high at the initial stage of protein synthesis, there is a possibility of only condensation and dehydration could proceed the process. Figure 4 shows that an amino acid glycine, which is drawn in pink, bonded with its carboxyl end to 3’-end of polyuridine at its bottom region of the left-handed single-stranded helix. This is going to polymerize with proline for further polymerization of L-amino acids. N-terminal end of glycine in pink is bonded to carboxyl end of proline in blue. This picture could be modified to show the participation of ATP. But, I did not think that it is prerequisite. This kind of stereo-specific chemical reaction could never have evolved on the outside surface in contact with hot water but might have been possible on the surface of silicates. If the left-handed RNA helix has a 6.5-fold symmetry, the stacking trouble of neighboring uracil bases does not occur. Figures 5 and 6 shows two stereo views of 6.5-fold symmetrical left-handed single-stranded polyuridine. In this case there is no need of worrying about the short contact between the nucleotides of two nearest neighbors, and the stacking base distance is also all right. Figure 7 shows that a peptide composed of L-amino acids having rather small side chains such as Ala, Ser, Val, Pro and Gly can be accommodated inside the cavity of the left-handed single-stranded helical sylinder, but those having large side chains could have a tendency to break the regularity.

Figure 1. Stereoview of three 6-fold left-handed single-stranded helices of 5’-phospho- D-ribose hydrogen bonded by U-U base pairs

Figure 2. Stereoview of the contact area of the three infinite helices intercalating one molecule of adenine triphosphate (ATP) which is drawn in green
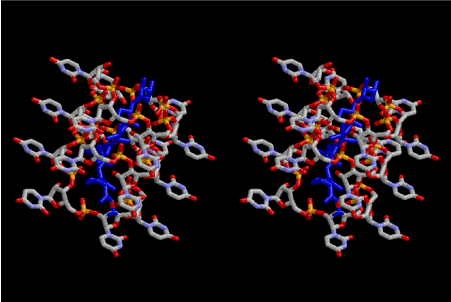
Figure 3. Stereoview of polymerized peptides, amino acid sequence of which are Gly-Pro- Gly-Asp-Ala-Ala-Ser-Val. The conformation of the polypeptide is not α–helix
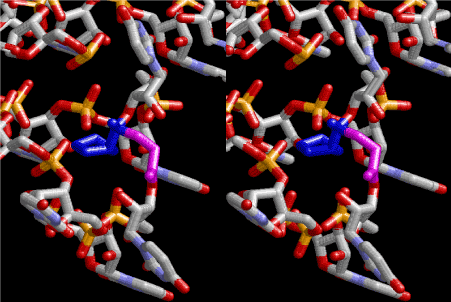
Figure 4. Stereoview of dimerization of glycine and proline. Glycine drawn in pink is bonded to 3’-end of polyuridine at its bottom region of the left-handed single-stranded helix. N-terminal end of proline in blue is bonded to carboxyl end of glycine
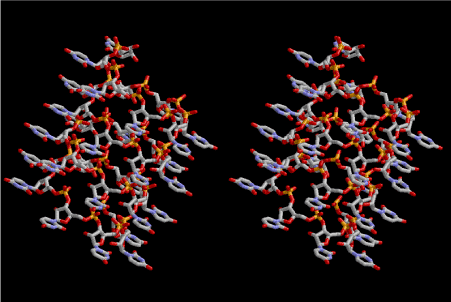
Figure 5. Stereoview of 6.5 fold symmetrical left-handed single-stranded polyuridine. This structure is also capable of base-pairing with the adjacent layer. The cavity in the layer is a little wider than that of 6-fold symmetrical one

Figure 6. This picture is the same as Fig. 5, but seen from a different direction in parallel with the axis of that of the left-handed helix
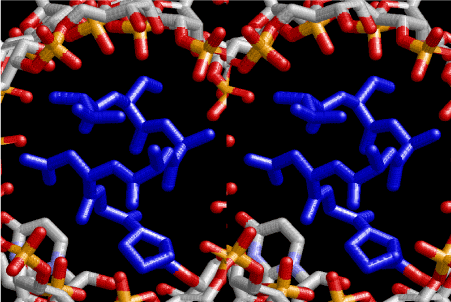
Figure 7. Stereoview of α–helical strucure of peptides drawn in blue and composed of L-amno acids, sequence of which is ProGlyAspAlaAlaSerVal
Accumulation of large L-amino acids in the cavity
When I published the basic idea about the accommodation of small amino acid polypeptide in the cavities of either 6-fold or 6.5-fold symmetry of left-handed RNA helices, I did not understand how the amino acids carrying large side-chains take over the world of ribosomal proteins. At that time, my hypothesis was the importance of interaction with the outside of the RNA layers, surrounded by silicate rocks. I think that the importance of silicate rocks still cannot be neglected, but here, I would like to propose another idea about the importance of 8-fold symmetry of left-handed RNA helices. Figure 8 shows a stereoview of the two left-handed helical forms of both 6-fold RNA below at the right-hand side, and the four left-handed 8-fold symmetrical lattices of RNA at the upper left-hand side. Two cavities are observed among the 6-fold lattices, while the four 8-fold lattices can make a bigger cavity that is suitable for accommodating amino acids having larger side-chains such as Arg, Met, Phe, Tyr, Trp etc. Figure 9 shows a most primitive shape of tRNA, which is formed from one turn of the 8-fold symmetrical left-handed RNA lattice. At its upper left-hand side, we can see the site of 3’-end, where amino acids are going to be bound by dehydration with a help of energy-supply from ATP. It is aminoacyl-tRNA synthesis. In this theory of aminoacyl-tRNA evolution, the aminoacylated tRNA just occupies the A-site of the most primitive ribosome. When the next aminoacylated tRNA comes up by splitting the neighboring ring of the 8-fold symmetrical RNA lattice, the former one automatically shifts to the neighboring site as the P-site tRNA. Peptidyl transfer occurs in the next place, and the deacylated tRNA leaves that site. Accordingly, aminoacylation and elongation began here cooperatively, although the role of messenger RNA (mRNA) does not come along it. Maybe, the role of mRNA would have come with the evolution of tRNA as formation of anticodon and anticodon loop.
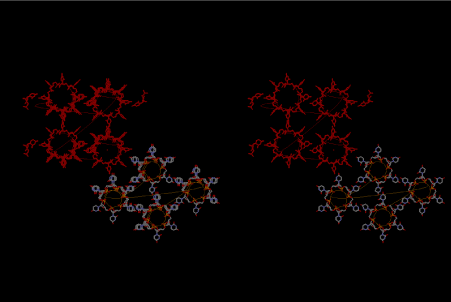
Figure 8. Stereoview of four 6-fold symmetrical left-handed single-stranded RNA helices at the bottom of right-hand corner, while four 8-fold one, which is drawn in red, are arranged at the upper left-hand side
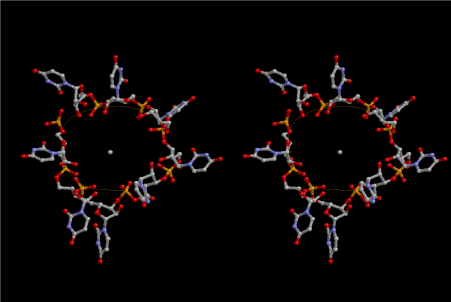
Figure 9. Most primitive form of tRNA composed of 8 uridines. This is made of four uracils at the 5’-end from the lower strand and four uracils at the 3’-end from the upper strand, and connected with each other. The 5’-end of uridine is base-paired with the corresponding one in the neighboring 8-fold lattice
Evolution of aminoacyl-tRNA synthetase
The evolution of aminoacyl-tRNA synthetase (aaRS) has led to two classes, Class I and Class II, based on structural sequence features of the catalytic core [3-5]. The evolution of Class I aaRS started from a dimeric form of RNA. The 8-fold symmetrical left-handed RNA is base-paired with another one by U-U hydrogen bonds. This primitive tRNA does not distinguish Class I and Class II. When tRNA evolves by adding anticodon and anticodon loop, those groups are favorable for interacting with the dimeric counterpart of the 8-fold symmetrical one. This is the beginning of the evolution of Class I aaRS. Figure 10 shows one primitive tRNA, which is drawn in Figure 9, is stacked on two rounds of 8-fold symmetrical left-handed single-stranded RNA helices. The primitive tRNA and the RNA are drawn in yellow. The 3’-end of the tRNA part is aminoacylated with arginine residue in blue, with a help of ATP at the other side of the 3’-end, which is the best position for dehydrating from aminoacyl group. By repeating this mechanism, the amino acids having larger side-chains such as Arg, Met, Phe, Tyr, and Trp etc are going to be accommodated. Disulfide bond formation by Cys residues from different 8-fold lattice could possibly be formed. Increase of Arg residues tends to make the cavity much larger and breaking the 8-fold lattice to bring about formation of right-handed double-stranded RNA helices, which are very popular in the field of the present-day biological chemistry.
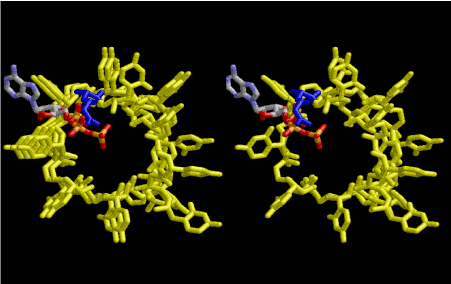
Figure 10. One primitive tRNA, which is drawn in Figure 9, is stacked on two rounds of 8-fold symmetrical left-handed single-stranded RNA helices. The primitive tRNA and the RNA are drawn in yellow. The 3’-end of the tRNA part is aminoacylated with arginine residue in blue, with a help of ATP at the other side of the 3’-end, which is the best position for dehydrating from aminoacyl group
On the other hand, the increase of amino acids having larger side-chains gives rise to an evolution of protein structure, which are composed of hydrophobic and hydrophilic groups. By this way, proteins are independently more important from the ribosomal proteins, and the highly complex world of the present-day living cells were produced by a step-wise change of RNA structure. Evolution of tRNA structure as a clover-leaf of primary structure and L-shaped tertiary structure was closely related with the development of the mechanism of aminoacylation.
References
- Nagano K (2017) Abiogenic evolution of ribonucleic acid as a left-handed helix and its protecting substances on primordial earth – A story about evolution of ribosome and the origine of life. Intergr Mol Med 4: 1-32.
- Joyce GF (1989) RNA evolution and the origins of life. Nature 338: 217-224. [Crossref]
- Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347: 203-206. [Crossref]
- Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature 347: 249-255. [Crossref]
- Ibba M, S?ll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617-650. [Crossref]










